Recursive partitioning for tumor classification with gene expression microarray data
- PMID: 11381113
- PMCID: PMC34421
- DOI: 10.1073/pnas.111153698
Recursive partitioning for tumor classification with gene expression microarray data
Abstract
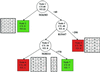
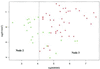
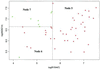
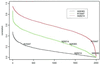
Precise classification of tumors is critically important for cancer diagnosis and treatment. It is also a scientifically challenging task. Recently, efforts have been made to use gene expression profiles to improve the precision of classification, with limited success. Using a published data set for purposes of comparison, we introduce a methodology based on classification trees and demonstrate that it is significantly more accurate for discriminating among distinct colon cancer tissues than other statistical approaches used heretofore. In addition, competing classification trees are displayed, which suggest that different genes may coregulate colon cancers.
Figures


References
-
- Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. - PubMed
-
- Stephenson J. J Am Med Assoc. 1999;282:927–928. - PubMed
-
- Theillet C. Nat Med. 1998;4:767–768. - PubMed
-
- Strausberg R L, Austin M J F. Physiol Genomics. 1999;1:25–32. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

