Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster
- PMID: 11381126
- PMCID: PMC34428
- DOI: 10.1073/pnas.121101598
Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster
Abstract
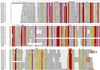
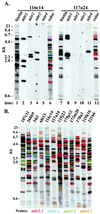
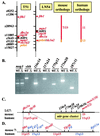
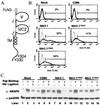
An extensive, highly diversified multigene family of novel immune-type receptor (nitr) genes has been defined in Danio rerio (zebrafish). The genes are predicted to encode type I transmembrane glycoproteins consisting of extracellular variable (V) and V-like C2 (V/C2) domains, a transmembrane region and a cytoplasmic tail. All of the genes examined encode immunoreceptor tyrosine-based inhibition motifs in the cytoplasmic tail. Radiation hybrid panel mapping and analysis of a deletion mutant line (b240) indicate that a minimum of approximately 40 nitr genes are contiguous in the genome and span approximately 0.6 Mb near the top of zebrafish linkage group 7. One flanking region of the nitr gene complex shares conserved synteny with a region of mouse chromosome 7, which shares conserved synteny with human 19q13.3-q13.4 that encodes the leukocyte receptor cluster. Antibody-induced crosslinking of Nitrs that have been introduced into a human natural killer cell line inhibits the phosphorylation of mitogen-activated protein kinase that is triggered by natural killer-sensitive tumor target cells. Nitrs likely represent intermediates in the evolution of the leukocyte receptor cluster.
Figures
References
-
- Medzhitov R, Janeway C A J. Cell. 1997;91:295–298. - PubMed
-
- Wende H, Colonna M, Ziegler A, Volz A. Mamm Genome. 1999;10:154–160. - PubMed
-
- Kubagawa H, Cooper M D, Chen C C, Ho L H, Alley T L, Hurez V, Tun T, Uehara T, Shimada T, Burrows P D. Curr Top Microbiol Immunol. 1999;244:137–149. - PubMed
-
- Moretta L, Biassoni R, Bottino C, Mingari M C, Moretta A. Immunol Today. 2000;21:420–422. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

