Preferential interaction of the core histone tail domains with linker DNA
- PMID: 11381129
- PMCID: PMC34399
- DOI: 10.1073/pnas.121171498
Preferential interaction of the core histone tail domains with linker DNA
Abstract
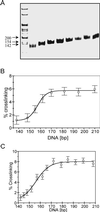
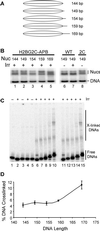
Within chromatin, the core histone tail domains play critical roles in regulating the structure and accessibility of nucleosomal DNA within the chromatin fiber. Thus, many nuclear processes are facilitated by concomitant posttranslational modification of these domains. However, elucidation of the mechanisms by which the tails mediate such processes awaits definition of tail interactions within chromatin. In this study we have investigated the primary DNA target of the majority of the tails in mononucleosomes. The results clearly show that the tails bind preferentially to "linker" DNA, outside of the DNA encompassed by the nucleosome core. These results have important implications for models of tail function within the chromatin fiber and for in vitro structural and functional studies using nucleosome core particles.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

