Localization of GAR transformylase in Escherichia coli and mammalian cells
- PMID: 11381136
- PMCID: PMC34393
- DOI: 10.1073/pnas.121182998
Localization of GAR transformylase in Escherichia coli and mammalian cells
Abstract
Enzymes of the de novo purine biosynthetic pathway may form a multienzyme complex to facilitate substrate flux through the ten serial steps constituting the pathway. One likely strategy for complex formation is the use of a structural scaffold such as the cytoskeletal network or subcellular membrane of the cell to mediate protein-protein interactions. To ascertain whether this strategy pertains to the de novo purine enzymes, the localization pattern of the third purine enzyme, glycinamide ribonucleotide transformylase (GAR Tfase) was monitored in live Escherichia coli and mammalian cells. Genes encoding human as well as E. coli GAR Tfase fused with green fluorescent protein (GFP) were introduced into their respective cells with regulated expression of proteins and localization patterns monitored by using confocal fluorescence microscopy. In both instances images showed proteins to be diffused throughout the cytoplasm. Thus, GAR Tfase is not localized to an existing cellular architecture, so this device is probably not used to concentrate the members of the pathway. However, discrete clusters of the pathway may still exist throughout the cytoplasm.
Figures
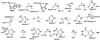

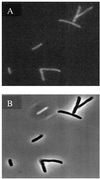



References
-
- Warren M S, Mattia K M, Marolewski A, Benkovic S J. Pure Appl Chem. 1996;68:2029–2036.
-
- Elion G B. Science. 1989;244:41. - PubMed
-
- Seeger D R, Cosulich D B, Smith J R, Hultquist M E. J Am Chem Soc. 1949;71:1753–1758.
-
- Inglese J, Johnson D L, Shiau A, Smith J M, Benkovic S J. Biochemistry. 1990;29:1436–1443. - PubMed
-
- Shim J H, Benkovic S J. Biochemistry. 1998;37:8776–8782. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

