A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast
- PMID: 11387218
- PMCID: PMC125490
- DOI: 10.1093/emboj/20.11.2857
A role for DNA polymerase alpha in epigenetic control of transcriptional silencing in fission yeast
Abstract
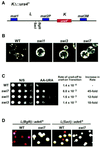
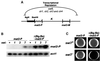
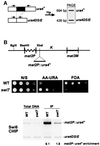
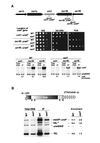
In the fission yeast Schizosaccharomyces pombe, transcriptional silencing at the mating-type region, centromeres and telomeres is epigenetically controlled, and results from the assembly of higher order chromatin structures. Chromatin proteins associated with these silenced loci are believed to serve as molecular bookmarks that help promote inheritance of the silenced state during cell division. Specifically, a chromodomain protein Swi6 is believed to be an important determinant of the epigenetic imprint. Here, we show that a mutation in DNA polymerase alpha (pol(alpha)) affects Swi6 localization at the mating-type region and causes a 45-fold increase in spontaneous transition from the silenced epigenetic state to the expressed state. We also demonstrate that pol(alpha) mutant cells are defective in Swi6 localization at centromeres and telomeres. Genetic analysis suggests that Polalpha and Swi6 are part of the same silencing pathway. Interestingly, we found that Swi6 directly binds to Pol(alpha) in vitro. Moreover, silencing-defective mutant Pol(alpha) displays reduced binding to Swi6 protein. This work indicates involvement of a DNA replication protein, Pol(alpha), in heterochromatin assembly and inheritance of epigenetic chromatin structures.
Figures


Similar articles
-
Chromodomain protein Swi6-mediated role of DNA polymerase alpha in establishment of silencing in fission Yeast.J Biol Chem. 2001 Dec 21;276(51):47814-21. doi: 10.1074/jbc.M109186200. Epub 2001 Oct 1. J Biol Chem. 2001. PMID: 11581276
-
Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals.Gene. 1994 May 27;143(1):139-43. doi: 10.1016/0378-1119(94)90619-x. Gene. 1994. PMID: 8200530
-
Transcriptional silencing in fission yeast.J Cell Physiol. 2000 Sep;184(3):311-8. doi: 10.1002/1097-4652(200009)184:3<311::AID-JCP4>3.0.CO;2-D. J Cell Physiol. 2000. PMID: 10911361 Review.
-
Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications.Genes Dev. 2002 Jul 15;16(14):1766-78. doi: 10.1101/gad.997702. Genes Dev. 2002. PMID: 12130537 Free PMC article.
-
Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe.Nucleic Acids Res. 2002 Apr 1;30(7):1465-82. doi: 10.1093/nar/30.7.1465. Nucleic Acids Res. 2002. PMID: 11917007 Free PMC article. Review.
Cited by
-
The replisome guides nucleosome assembly during DNA replication.Cell Biosci. 2020 Mar 12;10:37. doi: 10.1186/s13578-020-00398-z. eCollection 2020. Cell Biosci. 2020. PMID: 32190287 Free PMC article. Review.
-
The emerging role of SARS-CoV-2 nonstructural protein 1 (nsp1) in epigenetic regulation of host gene expression.FEMS Microbiol Rev. 2024 Sep 18;48(5):fuae023. doi: 10.1093/femsre/fuae023. FEMS Microbiol Rev. 2024. PMID: 39231808 Free PMC article. Review.
-
A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast.EMBO J. 2004 Oct 1;23(19):3825-35. doi: 10.1038/sj.emboj.7600401. Epub 2004 Sep 16. EMBO J. 2004. PMID: 15372076 Free PMC article.
-
Alterations of DNA and chromatin structures at telomeres and genetic instability in mouse cells defective in DNA polymerase alpha.Mol Cell Biol. 2005 Dec;25(24):11073-88. doi: 10.1128/MCB.25.24.11073-11088.2005. Mol Cell Biol. 2005. PMID: 16314528 Free PMC article.
-
The CINs of the centromere.Biochem Soc Trans. 2013 Dec;41(6):1706-11. doi: 10.1042/BST20130146. Biochem Soc Trans. 2013. PMID: 24256279 Free PMC article. Review.
References
-
- Allshire R.C. (1996) Transcriptional silencing in the fission yeast: a manifestation of higher order chromosome structure and function. In Russo,V.E.A., Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 443–466.
-
- Allshire R.C., Nimmo,E.R., Ekwall,K., Javerzat,J.P. and Cranston,G. (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev., 9, 218–233. - PubMed
-
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

