TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth
- PMID: 11390358
- PMCID: PMC312704
- DOI: 10.1101/gad.901101
TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth
Abstract
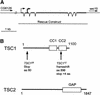
Tuberous sclerosis is a human disease caused by mutations in the TSC1 or the TSC2 tumor suppressor gene. Previous studies of a Drosophila TSC2 homolog suggested a role for the TSC genes in maintaining DNA content, with loss of TSC2 leading to polyploidy and increased cell size. We have isolated mutations in the Drosophila homolog of the TSC1 gene. We show that TSC1 and TSC2 form a complex and function in a common pathway to control cellular growth. Unlike previous studies, our work shows that TSC1(-) or TSC2(-) cells are diploid. We find that, strikingly, the heterozygosity of TSC1 or TSC2 is sufficient to rescue the lethality of loss-of-function insulin receptor mutants. Further genetic analyses suggest that the TSC genes act in a parallel pathway that converges on the insulin pathway downstream from Akt. Taken together, our studies identified the TSC tumor suppressors as novel negative regulators of insulin signaling.
Figures



References
-
- Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell. 1999;97:865–875. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
