Trypsin cleavage stabilizes the rotavirus VP4 spike
- PMID: 11390607
- PMCID: PMC114321
- DOI: 10.1128/JVI.75.13.6052-6061.2001
Trypsin cleavage stabilizes the rotavirus VP4 spike
Abstract
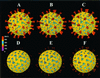
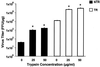
Trypsin enhances rotavirus infectivity by an unknown mechanism. To examine the structural basis of trypsin-enhanced infectivity in rotaviruses, SA11 4F triple-layered particles (TLPs) grown in the absence (nontrypsinized rotavirus [NTR]) or presence (trypsinized rotavirus [TR]) of trypsin were characterized to determine the structure, the protein composition, and the infectivity of the particles before and after trypsin treatment. As expected, VP4 was not cleaved in NTR particles and was cleaved into VP5(*) and VP8(*) in TR particles. However, surprisingly, while the VP4 spikes were clearly visible and well ordered in the electron cryomicroscopy reconstructions of TR TLPs, they were totally absent in the reconstructions of NTR TLPs. Biochemical analysis with radiolabeled particles indicated that the stoichiometry of the VP4 in NTR particles was the same as that in TR particles and that the VP8(*) portion of NTR, but not TR, particles is susceptible to further proteolysis by trypsin. Taken together, these structural and biochemical data show that the VP4 spikes in the NTR TLPs are icosahedrally disordered and that they are conformationally different. Structural studies on the NTR TLPs after trypsin treatment showed that spike structure could be partially recovered. Following additional trypsin treatment, infectivity was enhanced for both NTR and TR particles, but the infectivity of NTR remained 2 logs lower than that of TR particles. Increased infectivity in these particles corresponded to additional cleavages in VP5(*), at amino acids 259, 583, and putatively 467, which are conserved in all P serotypes of human and animal group A rotaviruses and also corresponded with a structural change in VP7. These biochemical and structural results show that trypsin cleavage imparts order to VP4 spikes on de novo synthesized virus particles, and these ordered spikes make virus entry into cells more efficient.
Figures

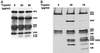



References
-
- Ball J M, Tian P, Zeng C Q Y, Morris A P, Estes M K. Age dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. - PubMed
-
- Beisner B, Kool D A, Marich A, Holmes I H. Characterization of G serotype dependent non-antibody inhibitors of rotavirus in normal mouse serum. Arch Virol. 1998;143:1277–1294. - PubMed
-
- Burns J W, Chen D, Estes M K, Ramig R F. Biological and immunological characterization of a simian rotavirus SA11 variant with an altered genome segment 4. Virology. 1989;169:427–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

