Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences
- PMID: 11390669
- PMCID: PMC87101
- DOI: 10.1128/MCB.21.13.4413-4424.2001
Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences
Abstract
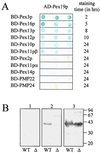
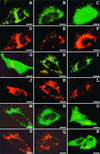
The molecular machinery underlying peroxisomal membrane biogenesis is not well understood. The observation that cells deficient in the peroxins Pex3p, Pex16p, and Pex19p lack peroxisomal membrane structures suggests that these molecules are involved in the initial stages of peroxisomal membrane formation. Pex19p, a predominantly cytosolic protein that can be farnesylated, binds multiple peroxisomal integral membrane proteins, and it has been suggested that it functions as a soluble receptor for the targeting of peroxisomal membrane proteins (PMPs) to the peroxisome. An alternative view proposes that Pex19p functions as a chaperone at the peroxisomal membrane. Here, we show that the peroxisomal sorting determinants and the Pex19p-binding domains of a number of PMPs are distinct entities. In addition, we extend the list of peroxins with which human Pex19p interacts to include the PMP Pex16p and show that Pex19p's CaaX prenylation motif is an important determinant in the affinity of Pex19p for Pex10p, Pex11pbeta, Pex12p, and Pex13p.
Figures






Similar articles
-
Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation.J Cell Sci. 2006 Sep 1;119(Pt 17):3539-50. doi: 10.1242/jcs.03100. Epub 2006 Aug 8. J Cell Sci. 2006. PMID: 16895967
-
Analysis of human Pex19p's domain structure by pentapeptide scanning mutagenesis.J Mol Biol. 2005 Mar 11;346(5):1275-86. doi: 10.1016/j.jmb.2005.01.013. Epub 2005 Jan 28. J Mol Biol. 2005. PMID: 15713480
-
The peroxisomal membrane protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway.J Cell Biol. 2008 Dec 29;183(7):1275-86. doi: 10.1083/jcb.200806062. J Cell Biol. 2008. PMID: 19114594 Free PMC article.
-
Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions.Biochim Biophys Acta. 2006 Dec;1763(12):1639-46. doi: 10.1016/j.bbamcr.2006.09.030. Epub 2006 Sep 26. Biochim Biophys Acta. 2006. PMID: 17069900 Review.
-
Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease.Biochim Biophys Acta. 2012 Sep;1822(9):1337-42. doi: 10.1016/j.bbadis.2012.06.004. Epub 2012 Jun 13. Biochim Biophys Acta. 2012. PMID: 22705440 Review.
Cited by
-
Conservation of PEX19-binding motifs required for protein targeting to mammalian peroxisomal and trypanosome glycosomal membranes.Eukaryot Cell. 2007 Aug;6(8):1439-49. doi: 10.1128/EC.00084-07. Epub 2007 Jun 22. Eukaryot Cell. 2007. PMID: 17586720 Free PMC article.
-
PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins.J Cell Biol. 2004 Mar 15;164(6):863-75. doi: 10.1083/jcb.200311131. Epub 2004 Mar 8. J Cell Biol. 2004. PMID: 15007061 Free PMC article.
-
Recognition and Chaperoning by Pex19, Followed by Trafficking and Membrane Insertion of the Peroxisome Proliferation Protein, Pex11.Cells. 2022 Jan 4;11(1):157. doi: 10.3390/cells11010157. Cells. 2022. PMID: 35011719 Free PMC article.
-
MoPex19, which is essential for maintenance of peroxisomal structure and woronin bodies, is required for metabolism and development in the rice blast fungus.PLoS One. 2014 Jan 14;9(1):e85252. doi: 10.1371/journal.pone.0085252. eCollection 2014. PLoS One. 2014. PMID: 24454828 Free PMC article.
-
PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins.J Cell Biol. 2004 Jan 5;164(1):57-67. doi: 10.1083/jcb.200304111. J Cell Biol. 2004. PMID: 14709540 Free PMC article.
References
-
- Amery L, Fransen M, De Nys K, Mannaerts G P, Van Veldhoven P P. Mitochondrial and peroxisomal targeting of 2-methylacyl-CoA racemase in human. J Lipid Res. 2000;41:1752–1759. - PubMed
-
- Baerends R J S, Faber K N, Kram A M, Kiel J A, van der Klei I J, Veenhuis M. A stretch of positively charged amino acids at the N-terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J Biol Chem. 2000;275:9986–9995. - PubMed
-
- Cadwell R C, Joyce G F. Mutagenic PCR. In: Dieffenbach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 583–585.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
