Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B
- PMID: 11390970
- PMCID: PMC34636
- DOI: 10.1073/pnas.121177598
Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B
Abstract
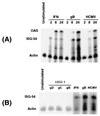
Human cytomegalovirus (HCMV) infection alters the expression of many cellular genes, including IFN-stimulated genes (ISGs) [Zhu, H., Cong, J.-P., Mamtora, G., Gingeras, T. & Shenk, T. (1998) Proc. Natl. Acad. Sci. USA 95, 14470-14475]. By using high-density cDNA microarrays, we show that the HCMV-regulated gene expression profile in fibroblasts does not differ substantially from the response generated by IFN. Furthermore, we identified the specific viral component triggering this response as the envelope glycoprotein B (gB). Cells treated with gB, but not other herpesviral glycoproteins, exhibited the same transcriptional profile as HCMV-infected cells. Thus, the interaction of gB with its as yet unidentified cellular receptor is the principal mechanism by which HCMV alters cellular gene expression early during infection. These findings highlight a pioneering paradigm for the consequences of virus-receptor interactions.
Figures



References
-
- Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. - PubMed
-
- Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P A. Science. 1996;274:610–614. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

