PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea
- PMID: 11391010
- PMCID: PMC34461
- DOI: 10.1073/pnas.111132998
PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea
Abstract
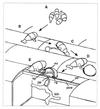
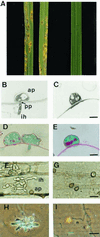
We describe in this study punchless, a nonpathogenic mutant from the rice blast fungus M. grisea, obtained by plasmid-mediated insertional mutagenesis. As do most fungal plant pathogens, M. grisea differentiates an infection structure specialized for host penetration called the appressorium. We show that punchless differentiates appressoria that fail to breach either the leaf epidermis or artificial membranes such as cellophane. Cytological analysis of punchless appressoria shows that they have a cellular structure, turgor, and glycogen content similar to those of wild type before penetration, but that they are unable to differentiate penetration pegs. The inactivated gene, PLS1, encodes a putative integral membrane protein of 225 aa (Pls1p). A functional Pls1p-green fluorescent protein fusion protein was detected only in appressoria and was localized in plasma membranes and vacuoles. Pls1p is structurally related to the tetraspanin family. In animals, these proteins are components of membrane signaling complexes controlling cell differentiation, motility, and adhesion. We conclude that PLS1 controls an appressorial function essential for the penetration of the fungus into host leaves.
Figures


Similar articles
-
Expression of Magnaporthe grisea avirulence gene ACE1 is connected to the initiation of appressorium-mediated penetration.Eukaryot Cell. 2007 Mar;6(3):546-54. doi: 10.1128/EC.00330-05. Epub 2006 Dec 1. Eukaryot Cell. 2007. PMID: 17142568 Free PMC article.
-
A multidrug resistance transporter in Magnaporthe is required for host penetration and for survival during oxidative stress.Plant Cell. 2006 Dec;18(12):3686-705. doi: 10.1105/tpc.105.037861. Epub 2006 Dec 22. Plant Cell. 2006. PMID: 17189344 Free PMC article.
-
Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus.Mol Microbiol. 2004 Sep;53(6):1695-707. doi: 10.1111/j.1365-2958.2004.04220.x. Mol Microbiol. 2004. PMID: 15341648
-
Cellular differentiation and host invasion by the rice blast fungus Magnaporthe grisea.Curr Opin Microbiol. 2007 Aug;10(4):339-45. doi: 10.1016/j.mib.2007.05.019. Epub 2007 Aug 17. Curr Opin Microbiol. 2007. PMID: 17707684 Review.
-
The role of glycerol in the pathogenic lifestyle of the rice blast fungus Magnaporthe oryzae.Environ Microbiol. 2017 Mar;19(3):1008-1016. doi: 10.1111/1462-2920.13688. Epub 2017 Mar 1. Environ Microbiol. 2017. PMID: 28165657 Review.
Cited by
-
Hyphopodium-Specific VdNoxB/VdPls1-Dependent ROS-Ca2+ Signaling Is Required for Plant Infection by Verticillium dahliae.PLoS Pathog. 2016 Jul 27;12(7):e1005793. doi: 10.1371/journal.ppat.1005793. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27463643 Free PMC article.
-
Morphogenesis in fungal pathogenicity: shape, size, and surface.PLoS Pathog. 2012;8(12):e1003027. doi: 10.1371/journal.ppat.1003027. Epub 2012 Dec 6. PLoS Pathog. 2012. PMID: 23236274 Free PMC article. No abstract available.
-
The bZIP transcription factor BIP1 of the rice blast fungus is essential for infection and regulates a specific set of appressorium genes.PLoS Pathog. 2024 Jan 22;20(1):e1011945. doi: 10.1371/journal.ppat.1011945. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38252628 Free PMC article.
-
Identification of virulence genes in Fusarium oxysporum f. sp. lycopersici by large-scale transposon tagging.Mol Plant Pathol. 2009 Jan;10(1):95-107. doi: 10.1111/j.1364-3703.2008.00512.x. Mol Plant Pathol. 2009. PMID: 19161356 Free PMC article.
-
Communication is key: extracellular vesicles as mediators of infection and defence during host-microbe interactions in animals and plants.FEMS Microbiol Rev. 2022 Jan 18;46(1):fuab044. doi: 10.1093/femsre/fuab044. FEMS Microbiol Rev. 2022. PMID: 34448857 Free PMC article. Review.
References
-
- Emmett R W, Parbery D G. Annu Rev Phytopathol. 1975;13:147–167.
-
- Dean R A. Annu Rev Phytopathol. 1997;35:211–234. - PubMed
-
- Hamer J E, Talbot N J. Curr Opin Microbiol. 1998;1:693–697. - PubMed
-
- Ou S H. Rice Diseases. 2nd Ed. Commonwealth Agricultural Bureaux, Kew, U.K.: Commonwealth Mycological Institute; 1985. pp. 109–201.
-
- Hamer J E, Howard R J, Chumley F G, Valent B. Science. 1988;239:288–290. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

