Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate
- PMID: 11395452
- PMCID: PMC95271
- DOI: 10.1128/JB.183.13.3890-3902.2001
Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate
Abstract
Escherichia coli responds to oxidative stress by activating sets of coregulated genes that help the cell to maintain homeostasis. Identified previously by genetic and biochemical approaches, the soxRS system mediates the induction of 18 of these redox-inducible genes (including the soxS gene itself). An overlapping set of genes is activated by an assortment of structurally unrelated molecules with antibiotic activities; many genes in this response are controlled by the marRAB system. The activation of either the soxRS or the marRAB system results in enhanced resistance to both superoxide-generating agents and multiple antibiotics. In order to probe the extent of these regulatory networks, we have measured whole-genome transcriptional profiles of the E. coli response to the superoxide-generating agent paraquat (PQ), an inducer of the soxRS system, and to the weak acid salt sodium salicylate (NaSal), an inducer of the marRA system. A total of 112 genes was modulated in response to PQ, while 134 genes were modulated in response to NaSal. We have also obtained transcriptional profiles of the SoxS and MarA regulons in the absence of global stress, in order to establish the regulatory hierarchies within the global responses. Several previously unrelated genes were shown to be under SoxS or MarA control. The genetic responses to both environmental insults revealed several common themes, including the activation of genes coding for functions that replenish reducing potential; regulate iron transport and storage; and participate in sugar and amino acid transport, detoxification, protein modification, osmotic protection, and peptidoglycan synthesis. A large number of PQ- and NaSal-responsive genes have no known function, suggesting that many adaptive metabolic changes that ensue after stress remain uncharacterized.
Figures
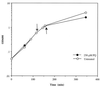


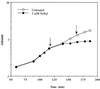

References
-
- Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem. 2000;275:29672–29684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

