Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster
- PMID: 11395469
- PMCID: PMC95288
- DOI: 10.1128/JB.183.13.4040-4051.2001
Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster
Abstract
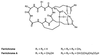
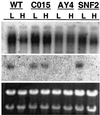
Ustilago maydis, the causal agent of corn smut disease, acquires and transports ferric ion by producing the extracellular, cyclic peptide, hydroxamate siderophores ferrichrome and ferrichrome A. Ferrichrome biosynthesis likely proceeds by hydroxylation and acetylation of L-ornithine, and later steps likely involve covalently bound thioester intermediates on a multimodular, nonribosomal peptide synthetase. sid1 encodes L-ornithine N(5)-oxygenase, which catalyzes hydroxylation of L-ornithine, the first committed step of ferrichrome and ferrichrome A biosynthesis in U. maydis. In this report we characterize sid2, another biosynthetic gene in the pathway, by gene complementation, gene replacement, DNA sequence, and Northern hybridization analysis. Nucleotide sequencing has revealed that sid2 is located 3.7 kb upstream of sid1 and encodes an intronless polypeptide of 3,947 amino acids with three iterated modules of an approximate length of 1,000 amino acids each. Multiple motifs characteristic of the nonribosomal peptide synthetase protein family were identified in each module. A corresponding iron-regulated sid2 transcript of 11 kb was detected by Northern hybridization analysis. By contrast, constitutive accumulation of this large transcript was observed in a mutant carrying a disruption of urbs1, a zinc finger, GATA family transcription factor previously shown to regulate siderophore biosynthesis in Ustilago. Multiple GATA motifs are present in the intergenic region between sid1 and sid2, suggesting bidirectional transcription regulation by urbs1 of this pathway. Indeed, mutation of two of these motifs, known to be important to regulation of sid1, altered the differential regulation of sid2 by iron.
Figures









References
-
- An Z, Farman M L, Taura S, Leong S A. New cosmid vectors for library construction, systematic chromosome walking and rapid restriction mapping in filamentous fungi. Gene. 1996;176:93–96. - PubMed
-
- Anke H, Anke T, Diekmann H. Biosynthesis of sideramines in fungi. Fusigen synthetase from extracts of Fusarium cubense. FEBS Lett. 1973;36:323–325. - PubMed
-
- Anke T, Diekmann H. Biosynthesis of sideramines in fungi. Rhodotorulic acid synthetase from extracts of Rhodotorula glutinis. FEBS Lett. 1972;27:259–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

