Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection
- PMID: 11401964
- PMCID: PMC98497
- DOI: 10.1128/IAI.69.7.4276-4286.2001
Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection
Abstract
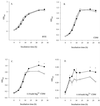
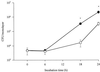
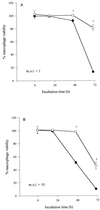
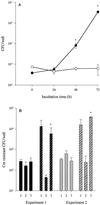
In the course of characterizing a locus involved in heme utilization, we identified a Legionella pneumophila gene predicted to encode a protein with homology to the product of the Salmonella enterica serovar Typhimurium pagP gene. In Salmonella, pagP increases resistance to the bactericidal effects of cationic antimicrobial peptides (CAMPs). Mutants with insertions in the L. pneumophila pagP-like gene were generated and showed decreased resistance to different structural classes of CAMPs compared to the wild type; hence, this gene was designated rcp for resistance to cationic antimicrobial peptides. Furthermore, Legionella CAMP resistance was induced by growth in low-magnesium medium. To determine whether rcp had any role in intracellular survival, mutants were tested in the two most relevant host cells for Legionnaires' disease, i.e., amoebae and macrophages. These mutants exhibited a 1,000-fold-decreased recovery during a Hartmannella vermiformis coculture. Complementation of the infectivity defect could be achieved by introduction of a plasmid containing the intact rcp gene. Mutations in rcp consistently reduced both the numbers of bacteria recovered during intracellular infection and their cytopathic capacity for U937 macrophages. The rcp mutant was also more defective for lung colonization of A/J mice. Growth of rcp mutants in buffered yeast extract broth was identical to that of the wild type, indicating that the observed differences in numbers of bacteria recovered from host cells were not due to a generalized growth defect. However, in low-Mg(2+) medium, the rcp mutant was impaired in stationary-phase survival. This is the first demonstration of a pagP-like gene, involved in resistance to CAMPs, being required for intracellular infection and virulence.
Figures


References
-
- Amemura-Maekawa J, Mishima-Abe S, Kura F, Takahashi T, Watanabe H. Identification of a novel periplasmic catalase-peroxidase KatA of Legionella pneumophila. FEMS Microbiol Lett. 1999;176:339–344. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

