Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(-)
- PMID: 11401985
- PMCID: PMC98518
- DOI: 10.1128/IAI.69.7.4447-4457.2001
Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(-)
Abstract
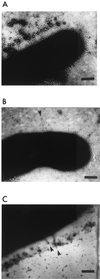
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:H(-) have emerged as important causes of diarrheal diseases and the hemolytic-uremic syndrome in Germany. In this study, we characterized a 32-kb fragment of the plasmid of SF EHEC O157:H(-), pSFO157, which differs markedly from plasmid pO157 of classical non-sorbitol-fermenting EHEC O157:H7. We found a cluster of six genes, termed sfpA, sfpH, sfpC, sfpD, sfpJ, and sfpG, which mediate mannose-resistant hemagglutination and the expression of fimbriae. sfp genes are similar to the pap genes, encoding P-fimbriae of uropathogenic E. coli, but the sfp cluster lacks homologues of genes encoding subunits of a tip fibrillum as well as regulatory genes. The major pilin, SfpA, despite its similarity to PapA, does not cluster together with known PapA alleles in a phylogenetic tree but is structurally related to the PmpA pilin of Proteus mirabilis. The putative adhesin gene sfpG, responsible for the hemagglutination phenotype, shows significant homology neither to papG nor to other known sequences. Sfp fimbriae are 3 to 5 nm in diameter, in contrast to P-fimbriae, which are 7 nm in diameter. PCR analyses showed that the sfp gene cluster is a characteristic of SF EHEC O157:H(-) strains and is not present in other EHEC isolates, diarrheagenic E. coli, or other Enterobacteriaceae. The sfp gene cluster is flanked by two blocks of insertion sequences and an origin of plasmid replication, indicating that horizontal gene transfer may have contributed to the presence of Sfp fimbriae in SF EHEC O157:H(-).
Figures






References
-
- Ammon A, Petersen L R, Karch H. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H−. J Infect Dis. 1999;179:1274–1277. - PubMed
-
- Baga M, Norgren M, Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49:241–251. - PubMed
-
- Bijlsma I G, Van Dijk L, Kusters J G, Gaastra W. Nucleotide sequences of two fimbrial major subunit genes, pmpA and ucaA, from canine-uropathogenic Proteus mirabilis strains. Microbiology. 1995;141:1349–1357. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

