HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres
- PMID: 11402064
- PMCID: PMC2192019
- DOI: 10.1083/jcb.153.6.1199
HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres
Abstract
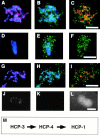
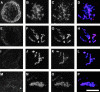
The centromere plays a critical role in the segregation of chromosomes during mitosis. In mammals, sister centromeres are resolved from one another in the G2 phase of the cell cycle. During prophase, chromosomes condense with sister centromeres oriented in a back to back configuration enabling only one chromatid to be captured by each half spindle. To study this process, we identified a centromere protein (CENP)-C-like protein, holocentric protein (HCP)-4, in Caenorhabditis elegans based on sequence identity, loss of function phenotype, and centromeric localization. HCP-4 is found in the cytoplasm during interphase, but is nuclear localized in mitosis, where it localizes specifically to the centromere. The localization of HCP-4 to the centromere is dependent on the centromeric histone HCP-3; in addition, HCP-3 and HCP-4 are both required for localization of a CENP-F-like protein, HCP-1, indicating an ordered assembly pathway. Loss of HCP-4 expression by RNA-mediated interference resulted in a failure to generate resolution of sister centromeres on chromosomes, suggesting that HCP-4 is required for sister centromere resolution. These chromosomes also failed to form a functional kinetochore. Thus, the CENP-C-like protein HCP-4 is essential for both resolution sister centromeres and attachment to the mitotic spindle.
Figures
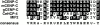





Comment in
-
Here, there, and everywhere: kinetochore function on holocentric chromosomes.J Cell Biol. 2001 Jun 11;153(6):F33-8. doi: 10.1083/jcb.153.6.f33. J Cell Biol. 2001. PMID: 11402076 Free PMC article. Review. No abstract available.
References
-
- Bailey T.L., Gribskov M. Combining evidence using p-valuesapplication to sequence homology searches. Bioinformatics. 1998;14:48–54. - PubMed
-
- Brinkley B.R., Stubblefield E. Ultrastructure and interaction of the kinetchore and centriole in mitosis and meiosis. In: Prescott D.M., Goldstein L., McConkey E., editors. Advances in Cell Biology. Vol. 1. Appleton-Century-Crofts; New York: 1970. pp. 119–185.
-
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone-H3-like protein in C. elegans . Nature. 1999;401:547–548. - PubMed
-
- Carrington W.A., Lynch R.M., Moore E.D., Isenberg G., Fogarty K.E., Fay F.S. Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science. 1995;268:1483–1487. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

