Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves
- PMID: 11402171
- PMCID: PMC135575
- DOI: 10.1105/tpc.13.6.1437
Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves
Abstract
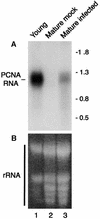
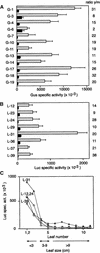
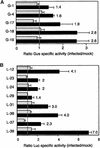
The geminivirus tomato golden mosaic virus (TGMV) amplifies its DNA genome in differentiated plant cells that lack detectable levels of DNA replication enzymes. Earlier studies showed that TGMV induces the accumulation of proliferating cell nuclear antigen (PCNA), the processivity factor for DNA polymerase delta, in mature cells of Nicotiana benthamiana. We sought to determine if PCNA protein accumulation reflects transcriptional activation of the host gene. RNA gel blot analysis detected an approximately 1200-nucleotide PCNA transcript in young leaves. The same RNA was found in mature leaves of infected but not healthy plants. Reporter gene analysis showed that a 633-bp promoter fragment of the N. benthamiana PCNA gene supports high levels of expression in cultured cells and in young but not mature leaves of healthy transgenic plants. In contrast, PCNA promoter activity was detected in both young and mature leaves of TGMV-infected plants. Developmental studies established a strong relationship between symptom severity, viral DNA accumulation, PCNA promoter activity, and endogenous PCNA mRNA levels. Mutation of an E2F consensus element in the PCNA promoter had no effect on its activity in young leaves but increased transcription in healthy mature leaves. Unlike the wild-type PCNA promoter, TGMV infection had no detectable effect on the activity of the mutant E2F promoter. Together, these results demonstrate that geminivirus infection induces the accumulation of a host replication factor by activating transcription of its gene in mature tissues, most likely by overcoming E2F-mediated repression.
Figures

References
-
- Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C., Helin, K., and Cella, R. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem. 275, 19258–19267. - PubMed
-
- Bass, H.W., Nagar, S., Hanley-Bowdoin, L., and Robertson, D. (2000). Chromosome condensation induced by geminivirus infection of mature plant cells. J. Cell Sci. 113, 1149–1160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

