The mitochondrial isovaleryl-coenzyme a dehydrogenase of arabidopsis oxidizes intermediates of leucine and valine catabolism
- PMID: 11402190
- PMCID: PMC111152
- DOI: 10.1104/pp.126.2.601
The mitochondrial isovaleryl-coenzyme a dehydrogenase of arabidopsis oxidizes intermediates of leucine and valine catabolism
Abstract
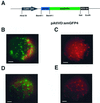
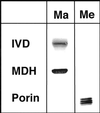
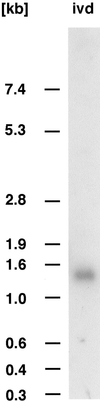
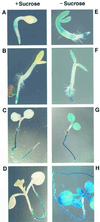
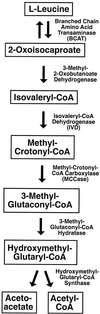
We recently identified a cDNA encoding a putative isovaleryl-coenzyme A (CoA) dehydrogenase in Arabidopsis (AtIVD). In animals, this homotetrameric enzyme is located in mitochondria and catalyzes the conversion of isovaleryl-CoA to 3-methylcrotonyl-CoA as an intermediate step in the leucine (Leu) catabolic pathway. Expression of AtIVD:smGFP4 fusion proteins in tobacco (Nicotiana tabacum) protoplasts and biochemical studies now demonstrate the in vivo import of the plant isovaleryl-CoA dehydrogenase (IVD) into mitochondria and the enzyme in the matrix of these organelles. Two-dimensional separation of mitochondrial proteins by blue native and SDS-PAGE and size determination of the native and overexpressed proteins suggest homodimers to be the dominant form of the plant IVD. Northern-blot hybridization and studies in transgenic Arabidopsis plants expressing Ativd promoter:gus constructs reveal strong expression of this gene in seedlings and young plants grown in the absence of sucrose, whereas promoter activity in almost all tissues is strongly inhibited by exogeneously added sucrose. Substrate specificity tests with AtIVD expressed in Escherichia coli indicate a strong preference toward isovaleryl-CoA but surprisingly also show considerable activity with isobutyryl-CoA. This strongly indicates a commitment of the enzyme in Leu catabolism, but the activity observed with isobutyryl-CoA also suggests a parallel involvement of the enzyme in the dehydrogenation of intermediates of the valine degradation pathway. Such a dual activity has not been observed with the animal IVD and may suggest a novel connection of the Leu and valine catabolism in plants.
Figures
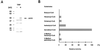

References
-
- Aubert S, Alban C, Bligny R, Douce R. Induction of β-methylcrotonyl-coenzyme A carboxylase in higher plant cells during carbohydrate starvation: evidence for a role of MCCase in leucine catabolism. FEBS Lett. 1996;383:175–180. - PubMed
-
- Bechtold N, Ellis J, Peletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci. 1993;316:1194–1199.
-
- Binder S, Brennicke A. Transcription initiation sites in mitochondria of Oenothera berteriana. J Biol Chem. 1993;268:7849–7855. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

