Inventory of the superfamily of P-type ion pumps in Arabidopsis
- PMID: 11402198
- PMCID: PMC111160
- DOI: 10.1104/pp.126.2.696
Inventory of the superfamily of P-type ion pumps in Arabidopsis
Abstract
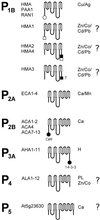
A total of 45 genes encoding for P-type ATPases have been identified in the complete genome sequence of Arabidopsis. Thus, this plant harbors a primary transport capability not seen in any other eukaryotic organism sequenced so far. The sequences group in all five subfamilies of P-type ATPases. The most prominent subfamilies are P(1B) ATPases (heavy metal pumps; seven members), P(2A) and P(2B) ATPases (Ca(2+) pumps; 14 in total), P(3A) ATPases (plasma membrane H(+) pumps; 12 members including a truncated pump, which might represent a pseudogene or an ATPase-like protein with an alternative function), and P(4) ATPases (12 members). P(4) ATPases have been implicated in aminophosholipid flipping but it is not known whether this is a direct or an indirect effect of pump activity. Despite this apparent plethora of pumps, Arabidopsis appears to be lacking Na(+) pumps and secretory pathway (PMR1-like) Ca(2+)-ATPases. A cluster of Arabidopsis heavy metal pumps resembles bacterial Zn(2+)/Co(2+)/Cd(2+)/Pb(2+) transporters. Two members of the cluster have extended C termini containing putative heavy metal binding motifs. The complete inventory of P-type ATPases in Arabidopsis is an important starting point for reverse genetic and physiological approaches aiming at elucidating the biological significance of these pumps.
Figures

References
-
- Allen GJ, Chu SP, Schumacher K, Shimazaki CT, Vafeados D, Kemper A, Hawke SD, Tallman G, Tsien RY, Harper JF. Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science. 2000;289:2338–2342. - PubMed
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. - PubMed
-
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. - PubMed
-
- Beard SJ, Hashim R, Membrillo-Hernandez J, Hughes MN, Poole RK. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25:883–891. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

