Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling
- PMID: 11402208
- PMCID: PMC111170
- DOI: 10.1104/pp.126.2.801
Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling
Abstract
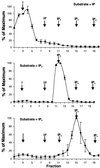
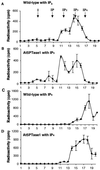
The inositol triphosphate (IP(3))-signaling pathway has been associated with several developmental and physiological processes in plants, but we currently know little about the regulation of this pathway. Inositol 5' phosphatases (5PTases) are enzymes that remove a 5' phosphate from several potential second messengers, including IP(3). In catalyzing the removal of a 5' phosphate from second messenger substrates, 5PTases can act to terminate signal transduction events. We describe the molecular analysis of At5PTase1, a 5PTase gene from Arabidopsis. When expressed transiently in Arabidopsis leaf tissue or ectopically in transgenic plants, At5PTase1 allowed for the increased hydrolysis of I(1,4,5)P(3) and I(1,3,4,5)P(4) substrates. At5PTase1 did not hydrolyze I(1)P, I(1,4)P(2), or PI(4,5)P(2) substrates. This substrate specificity was similar to that of the human Type I 5PTase. We identified 14 other potential At5PTase genes and constructed an unrooted phylogenetic tree containing putative Arabidopsis, human, and yeast 5PTase proteins. This analysis indicated that the Arabidopsis 5PTases were grouped in two separate branches of the tree. The multiplicity of At5PTases indicates that these enzymes may have different substrate specificities and play different roles in signal termination in Arabidopsis.
Figures
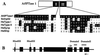



References
-
- Berridge MJ. Inositol trisphosphate and calcium signaling. Nature. 1993;361:315–325. - PubMed
-
- Chandra S, Low PS. Measurement of Ca2+fluxes during elicitation of the oxidative burst in aequorin-transformed tobacco cells. J Biol Chem. 1997;272:28274–28280. - PubMed
-
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. - PubMed
-
- Communi D, Lecocq R, Erneux C. Arginine 343 and 350 are two active residues involved in substrate binding by human Type I D-myo-inositol 1,4,5,-trisphosphate 5-phosphatase. J Biol Chem. 1996;271:11676–11683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

