Repellent signaling by Slit requires the leucine-rich repeats
- PMID: 11404414
- PMCID: PMC6762740
- DOI: 10.1523/JNEUROSCI.21-12-04290.2001
Repellent signaling by Slit requires the leucine-rich repeats
Abstract
Slit is a repellent axon guidance cue produced by the midline glia in Drosophila that is required to regulate the formation of contralateral projections and the lateral position of longitudinal tracts. Four sequence motifs comprise the structure of Slit: a leucine-rich repeat (LRR), epidermal growth factor-like (EGF) repeats, a laminin-like globular (G)-domain, and a cysteine domain. Here we demonstrate that the LRR is required for repellent signaling and in vitro binding to Robo. Repellent signaling by slit is reduced by point mutations that encode single amino acid changes in the LRR domain. By contrast to the EGF or G-domains, the LRR domain is required in transgenes to affect axon guidance. Finally, we show that the midline repellent receptor, Robo, binds Slit proteins with internal deletions that also retain repellent activity. However, Robo does not bind Slit protein missing the LRR. Taken together, our data demonstrate that Robo binding and repellent signaling by Slit require the LRR region.
Figures
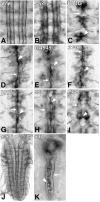
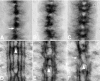
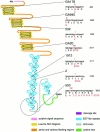

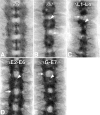

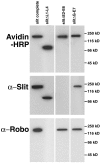
References
-
- Battye R, Stevens A, Jacobs JR. Axon repulsion from the midline of the Drosophila CNS requires slit function. Development. 1999;126:2475–2481. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Brose K, Bland KS, Wang K-H, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Evolutionary conservation of the repulsive guidance function of Slit proteins and their interactions with robo receptors. Cell. 1999;96:795–806. - PubMed
-
- Cornish T, Chi J, Johnson S, Lu Y, Campanelli JT. Globular domains of agrin are functional units that collaborate to induce acetylcholine receptor clustering. J Cell Sci. 1999;112:1213–1223. - PubMed
-
- Gay NJ, Packman LC, Weldon MA, Barna JC. A leucine-rich repeat peptide derived from the Drosophila Toll receptor forms extended filaments with a beta-sheet structure. FEBS Lett. 1991;291:87–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
