Exploring the DNA-binding specificities of zinc fingers with DNA microarrays
- PMID: 11404456
- PMCID: PMC34639
- DOI: 10.1073/pnas.111163698
Exploring the DNA-binding specificities of zinc fingers with DNA microarrays
Abstract
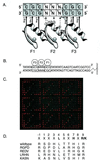
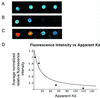
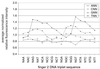
A key step in the regulation of networks that control gene expression is the sequence-specific binding of transcription factors to their DNA recognition sites. A more complete understanding of these DNA-protein interactions will permit a more comprehensive and quantitative mapping of the regulatory pathways within cells, as well as a deeper understanding of the potential functions of individual genes regulated by newly identified DNA-binding sites. Here we describe a DNA microarray-based method to characterize sequence-specific DNA recognition by zinc-finger proteins. A phage display library, prepared by randomizing critical amino acid residues in the second of three fingers of the mouse Zif268 domain, provided a rich source of zinc-finger proteins with variant DNA-binding specificities. Microarrays containing all possible 3-bp binding sites for the variable zinc fingers permitted the quantitation of the binding site preferences of the entire library, pools of zinc fingers corresponding to different rounds of selection from this library, as well as individual Zif268 variants that were isolated from the library by using specific DNA sequences. The results demonstrate the feasibility of using DNA microarrays for genome-wide identification of putative transcription factor-binding sites.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

