Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine
- PMID: 11406586
- PMCID: PMC150209
- DOI: 10.1093/emboj/20.12.3092
Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine
Abstract
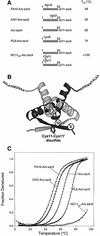
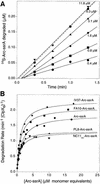
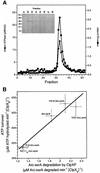
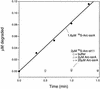
ClpXP is an ATP-dependent protease that denatures native proteins and translocates the denatured polypeptide into an interior peptidase chamber for degradation. To address the mechanism of these processes, Arc repressor variants with dramatically different stabilities and unfolding half-lives varying from months to seconds were targeted to ClpXP by addition of the ssrA degradation tag. Remarkably, ClpXP degraded each variant at a very similar rate and hydrolyzed approximately 150 molecules of ATP for each molecule of substrate degraded. The hyperstable substrates did, however, slow the ClpXP ATPase cycle. These results confirm that ClpXP uses an active mechanism to denature its substrates, probably one that applies mechanical force to the native structure. Furthermore, the data suggest that denaturation is inherently inefficient or that significant levels of ATP hydrolysis are required for other reaction steps. ClpXP degraded disulfide-cross-linked dimers efficiently, even when just one subunit contained an ssrA tag. This result indicates that the pore through which denatured proteins enter the proteolytic chamber must be large enough to accommodate simultaneous passage of two or three polypeptide chains.
Figures



References
-
- Bowie J.U. and Sauer,R.T. (1989) Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry, 28, 7139–7143. - PubMed
-
- Breg J.N., van Opheusden,J.H., Burgering,M.J., Boelens,R. and Kaptein,R. (1990) Structure of Arc repressor in solution: evidence for a family of β-sheet DNA-binding proteins. Nature, 346, 586–589. - PubMed
-
- Burgering M.J.M., Hald,M., Boelens,R., Breg,J.N. and Kaptein,R. (1995) Hydrogen exchange studies of the Arc repressor: evidence for a monomeric folding intermediate. Biopolymers, 35, 217–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

