Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection
- PMID: 11406603
- PMCID: PMC150207
- DOI: 10.1093/emboj/20.12.3272
Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection
Abstract
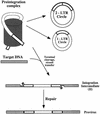
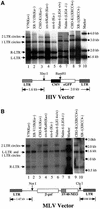
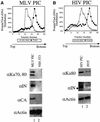
Early after infection, the retroviral RNA genome is reverse transcribed to generate a linear cDNA copy, then that copy is integrated into a chromosome of the host cell. We report that unintegrated viral cDNA is a substrate for the host cell non-homologous DNA end joining (NHEJ) pathway, which normally repairs cellular double-strand breaks by end ligation. NHEJ activity was found to be required for an end-ligation reaction that circularizes a portion of the unintegrated viral cDNA in infected cells. Consistent with this, the NHEJ proteins Ku70 and Ku80 were found to be bound to purified retroviral replication intermediates. Cells defective in NHEJ are known to undergo apoptosis in response to retroviral infection, a response that we show requires reverse transcription to form the cDNA genome but not subsequent integration. We propose that the double-strand ends present in unintegrated cDNA promote apoptosis, as is known to be the case for chromosomal double-strand breaks, and cDNA circularization removes the pro-apoptotic signal.
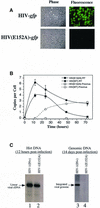
Figures

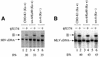


References
-
- Arad U. (1998) Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques, 24, 760–762. - PubMed
-
- Beall E.L. and Rio,D.C. (1996) Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev., 10, 921–933. - PubMed
-
- Bowerman B., Brown,P.O., Bishop,J.M. and Varmus,H.E. (1989) A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev., 3, 469–478. - PubMed
-
- Brown P.O., Bowerman,B., Varmus,H.E. and Bishop,J.M. (1987) Correct integration of retroviral DNA in vitro. Cell, 49, 347–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

