Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus
- PMID: 11408208
- PMCID: PMC90585
- DOI: 10.1128/AAC.45.7.1955-1963.2001
Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus
Abstract
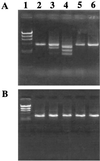
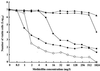
We report on the structural diversity of mecA gene complexes carried by 38 methicillin-resistant Staphylococcus aureus and 91 methicillin-resistant coagulase-negative Staphylococcus strains of seven different species with a special reference to its correlation with phenotypic expression of methicillin resistance. The most prevalent and widely disseminated mec complex had the structure mecI-mecR1-mecA-IS431R (or IS431mec), designated the class A mecA gene complex. In contrast, in S. haemolyticus, mecA was bracketed by two copies of IS431, forming the structure IS431L-mecA-IS431R. Of the 38 S. haemolyticus strains, 5 had low-level methicillin resistance (MIC, 1 to 4 mg/liter) and characteristic heterogeneous methicillin resistance as judged by population analysis. In these five strains, IS431L was located to the left of an intact mecI gene, forming the structure IS431L-class A mecA-gene complex. In other S. haemolyticus strains, IS431L was associated with the deletion of mecI and mecR1, forming the structure IS431L-DeltamecR1-mecA-IS431mec, designated the class C mecA gene complex. Mutants with the class C mecA gene complex were obtained in vitro by selecting strain SH621, containing the IS431L-class A mecA gene complex with low concentrations of methicillin (1 and 3 mg/liter). The mutants had intermediate level of methicillin resistance (MIC, 16 to 64 mg/liter). The mecA gene transcription was shown to be derepressed in a representative mutant strain, SH621-37. Our study indicated that the mecI-encoded repressor function is responsible for the low-level methicillin resistance of some S. haemolyticus clinical strains and that the IS431-mediated mecI gene deletion causes the expression of methicillin resistance through the derepression of mecA gene transcription.
Figures



Similar articles
-
Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus.J Bacteriol. 1996 Sep;178(18):5464-71. doi: 10.1128/jb.178.18.5464-5471.1996. J Bacteriol. 1996. PMID: 8808937 Free PMC article.
-
Analysis of mec regulator genes in clinical methicillin-resistant Staphylococcus aureus isolates according to the production of coagulase, types of enterotoxin, and toxic shock syndrome toxin-1.Hiroshima J Med Sci. 1999 Jun;48(2):49-56. Hiroshima J Med Sci. 1999. PMID: 10434474
-
Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control.Antimicrob Agents Chemother. 2001 Mar;45(3):815-24. doi: 10.1128/AAC.45.3.815-824.2001. Antimicrob Agents Chemother. 2001. PMID: 11181367 Free PMC article.
-
Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications.Clin Microbiol Rev. 1997 Oct;10(4):781-91. doi: 10.1128/CMR.10.4.781. Clin Microbiol Rev. 1997. PMID: 9336672 Free PMC article. Review.
-
Molecular genetics of methicillin-resistant Staphylococcus aureus.Int J Med Microbiol. 2002 Jul;292(2):67-74. doi: 10.1078/1438-4221-00192. Int J Med Microbiol. 2002. PMID: 12195737 Review.
Cited by
-
Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI.Antimicrob Agents Chemother. 2006 Oct;50(10):3457-9. doi: 10.1128/AAC.00629-06. Antimicrob Agents Chemother. 2006. PMID: 17005831 Free PMC article.
-
Morphological and genetic differences in two isogenic Staphylococcus aureus strains with decreased susceptibilities to vancomycin.Antimicrob Agents Chemother. 2003 Feb;47(2):568-76. doi: 10.1128/AAC.47.2.568-576.2003. Antimicrob Agents Chemother. 2003. PMID: 12543661 Free PMC article.
-
Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children.J Clin Microbiol. 2008 Nov;46(11):3778-83. doi: 10.1128/JCM.02262-07. Epub 2008 Oct 1. J Clin Microbiol. 2008. PMID: 18832123 Free PMC article.
-
Typing of staphylococcal cassette chromosome mec encoding methicillin resistance in Staphylococcus aureus strains isolated at the bone marrow transplant centre of Tunisia.Curr Microbiol. 2009 Oct;59(4):380-5. doi: 10.1007/s00284-009-9448-1. Epub 2009 Jul 9. Curr Microbiol. 2009. PMID: 19588195
-
Adjunctive mecA PCR for routine detection of methicillin susceptibility in clinical isolates of coagulase-negative staphylococci.J Clin Microbiol. 2014 May;52(5):1678-81. doi: 10.1128/JCM.02834-13. Epub 2014 Mar 12. J Clin Microbiol. 2014. PMID: 24622101 Free PMC article.
References
-
- Barberis-Maino L, Berger-Bächi B, Weber H, Beck W D, Kayser F H. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59:107–113. - PubMed
-
- Byrne M E, Gillespie M T, Skurray R A. 4′,4"-Adenyltransferase activity on conjugative plasmids isolated from Staphylococcus aureus is encoded on an integrated copy of pUB110. Plasmid. 1991;25:70–75. - PubMed
-
- Byrne M E, Littlejohn T G, Skurray R A. Transposons and insertion sequences in the evolution of multiresistant Staphylococcus aureus. In: Novick R P, editor. Molecular biology of staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 1303–1311.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

