Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87)
- PMID: 11408214
- PMCID: PMC90591
- DOI: 10.1128/AAC.45.7.1994-2000.2001
Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87)
Abstract
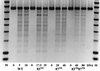
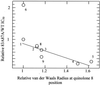
DNA gyrase is a target of quinolone antibacterial agents, but the molecular details of the quinolone-gyrase interaction are not clear. Quinolone resistance mutations frequently occur at residues Ser(83) and Asp(87) of the gyrase A subunit, suggesting that these residues are involved in drug binding. Single and double alanine substitutions were created at these positions (Ala(83), Ala(87), and Ala(83) Ala(87)), and the mutant proteins were assessed for DNA supercoiling, DNA cleavage, and resistance to a number of quinolone drugs. The Ala(83) mutant was fully active in supercoiling, whereas the Ala(87) and the double mutant were 2.5- and 4- to 5-fold less active, respectively; this loss in activity may be partly due to an increased affinity of these mutant proteins for DNA. Supercoiling inhibition and cleavage assays revealed that the double mutant has a high level of resistance to certain quinolones while the mutants with single alanine substitutions show low-level resistance. Using a drug-binding assay we demonstrated that the double-mutant enzyme-DNA complex has a lower affinity for ciprofloxacin than the wild-type complex. Based on the pattern of resistance to a series of quinolones, an interaction between the C-8 group of the quinolone and the double-mutant gyrase in the region of residues 83 and 87 is proposed.
Figures




References
-
- Bates A D, Maxwell A. DNA topology. Oxford, United Kingdom: IRL Press; 1993.
-
- Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure at 2.7 Å resolution of a 92K yeast DNA topoisomerase II fragment. Nature. 1996;379:225–232. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Caron P R. Appendix: compendium of DNA topoisomerase sequences. In: Bjornsti M-A, Osheroff N, editors. DNA topoisomerase protocols. DNA topology and enzymes. Vol. 94. Totowa, N.J: Humana Press; 1999. pp. 279–316. - PubMed
-
- Chen C-R, Malik M, Snyder M, Drlica M. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

