Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle
- PMID: 11408575
- PMCID: PMC37331
- DOI: 10.1091/mbc.12.6.1645
Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle
Abstract
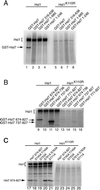
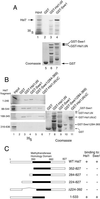
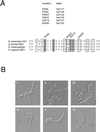
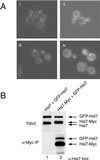
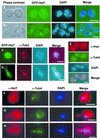
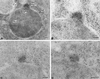
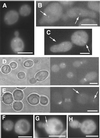
In Saccharomyces cerevisiae, entry into mitosis requires activation of the cyclin-dependent kinase Cdc28 in its cyclin B (Clb)-associated form. Clb-bound Cdc28 is susceptible to inhibitory tyrosine phosphorylation by Swe1 protein kinase. Swe1 is itself negatively regulated by Hsl1, a Nim1-related protein kinase, and by Hsl7, a presumptive protein-arginine methyltransferase. In vivo all three proteins localize to the bud neck in a septin-dependent manner, consistent with our previous proposal that formation of Hsl1-Hsl7-Swe1 complexes constitutes a checkpoint that monitors septin assembly. We show here that Hsl7 is phosphorylated by Hsl1 in immune-complex kinase assays and can physically associate in vitro with either Hsl1 or Swe1 in the absence of any other yeast proteins. With the use of both the two-hybrid method and in vitro binding assays, we found that Hsl7 contains distinct binding sites for Hsl1 and Swe1. A differential interaction trap approach was used to isolate four single-site substitution mutations in Hsl7, which cluster within a discrete region of its N-terminal domain, that are specifically defective in binding Hsl1. When expressed in hsl7Delta cells, each of these Hsl7 point mutants is unable to localize at the bud neck and cannot mediate down-regulation of Swe1, but retains other functions of Hsl7, including oligomerization and association with Swe1. GFP-fusions of these Hsl1-binding defective Hsl7 proteins localize as a bright perinuclear dot, but never localize to the bud neck; likewise, in hsl1Delta cells, a GFP-fusion to wild-type Hsl7 or native Hsl7 localizes to this dot. Cell synchronization studies showed that, normally, Hsl7 localizes to the dot, but only in cells in the G1 phase of the cell cycle. Immunofluorescence analysis and immunoelectron microscopy established that the dot corresponds to the outer plaque of the spindle pole body (SPB). These data demonstrate that association between Hsl1 and Hsl7 at the bud neck is required to alleviate Swe1-imposed G2-M delay. Hsl7 localization at the SPB during G1 may play some additional role in fine-tuning the coordination between nuclear and cortical events before mitosis.
Figures


Similar articles
-
Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae.Mol Cell Biol. 1999 Oct;19(10):7123-37. doi: 10.1128/MCB.19.10.7123. Mol Cell Biol. 1999. PMID: 10490648 Free PMC article.
-
Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast.Genes Dev. 1999 Jan 15;13(2):176-87. doi: 10.1101/gad.13.2.176. Genes Dev. 1999. PMID: 9925642 Free PMC article.
-
Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast.EMBO J. 2005 Jun 15;24(12):2194-204. doi: 10.1038/sj.emboj.7600683. Epub 2005 May 26. EMBO J. 2005. PMID: 15920482 Free PMC article.
-
Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae.Curr Opin Genet Dev. 2000 Feb;10(1):47-53. doi: 10.1016/s0959-437x(99)00051-9. Curr Opin Genet Dev. 2000. PMID: 10679396 Review.
-
Orchestrating the cell cycle in yeast: sequential localization of key mitotic regulators at the spindle pole and the bud neck.Microbiology (Reading). 2002 Sep;148(Pt 9):2647-2659. doi: 10.1099/00221287-148-9-2647. Microbiology (Reading). 2002. PMID: 12213912 Review. No abstract available.
Cited by
-
Determinants of Swe1p degradation in Saccharomyces cerevisiae.Mol Biol Cell. 2002 Oct;13(10):3560-75. doi: 10.1091/mbc.e02-05-0283. Mol Biol Cell. 2002. PMID: 12388757 Free PMC article.
-
Hitchhiking and epistasis give rise to cohort dynamics in adapting populations.Proc Natl Acad Sci U S A. 2017 Aug 1;114(31):8330-8335. doi: 10.1073/pnas.1702314114. Epub 2017 Jul 18. Proc Natl Acad Sci U S A. 2017. PMID: 28720700 Free PMC article.
-
A Synthetic Dosage Lethal Genetic Interaction Between CKS1B and PLK1 Is Conserved in Yeast and Human Cancer Cells.Genetics. 2016 Oct;204(2):807-819. doi: 10.1534/genetics.116.190231. Epub 2016 Aug 24. Genetics. 2016. PMID: 27558135 Free PMC article.
-
Cell cycle checkpoint regulators reach a zillion.Cell Cycle. 2013 May 15;12(10):1501-9. doi: 10.4161/cc.24637. Epub 2013 Apr 17. Cell Cycle. 2013. PMID: 23598718 Free PMC article. Review.
-
Reconstitution of the mammalian PI3K/PTEN/Akt pathway in yeast.Biochem J. 2005 Sep 1;390(Pt 2):613-23. doi: 10.1042/BJ20050574. Biochem J. 2005. PMID: 15913452 Free PMC article.
References
-
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. - PubMed
-
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. - PubMed
-
- Balasubramanian MK, McCollum D, Surana U. Tying the knot: linking cytokinesis to the nuclear cycle. J Cell Sci. 2000;113:1503–1513. - PubMed
-
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

