Identification of a Saccharomyces cerevisiae gene that is required for G1 arrest in response to the lipid oxidation product linoleic acid hydroperoxide
- PMID: 11408586
- PMCID: PMC37342
- DOI: 10.1091/mbc.12.6.1801
Identification of a Saccharomyces cerevisiae gene that is required for G1 arrest in response to the lipid oxidation product linoleic acid hydroperoxide
Abstract
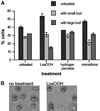
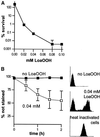
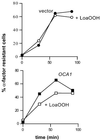
Reactive oxygen species cause damage to all of the major cellular constituents, including peroxidation of lipids. Previous studies have revealed that oxidative stress, including exposure to oxidation products, affects the progression of cells through the cell division cycle. This study examined the effect of linoleic acid hydroperoxide, a lipid peroxidation product, on the yeast cell cycle. Treatment with this peroxide led to accumulation of unbudded cells in asynchronous populations, together with a budding and replication delay in synchronous ones. This observed modulation of G1 progression could be distinguished from the lethal effects of the treatment and may have been due to a checkpoint mechanism, analogous to that known to be involved in effecting cell cycle arrest in response to DNA damage. By examining several mutants sensitive to linoleic acid hydroperoxide, the YNL099c open reading frame was found to be required for the arrest. This gene (designated OCA1) encodes a putative protein tyrosine phosphatase of previously unknown function. Cells lacking OCA1 did not accumulate in G1 on treatment with linoleic acid hydroperoxide, nor did they show a budding, replication, or Start delay in synchronous cultures. Although not essential for adaptation or immediate cellular survival, OCA1 was required for growth in the presence of linoleic acid hydroperoxide, thus indicating that it may function in linking growth, stress responses, and the cell cycle. Identification of OCA1 establishes cell cycle arrest as an actively regulated response to oxidative stress and will enable further elucidation of oxidative stress-responsive signaling pathways in yeast.
Figures


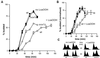

References
-
- Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of protein-tyrosine phosphatase 1B. J Biol Chem. 1999;274:34543–34546. - PubMed
-
- Binley KM, Radcliffe PA, Trevethick J, Duffy KA, Sudbery PE. The yeast PRS3 gene is required for cell integrity, cell cycle arrest upon nutrient deprivation, ion homeostasis, and the proper organization of the actin cytoskeleton. Yeast. 1999;15:1459–1469. - PubMed
-
- Breeden LL. Alpha-factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–342. - PubMed
-
- Carballo M, Conde M, El Bekay R, Martin-Nieto J, Camacho MJ, Monteseirin J, Conde J, Bedoya FJ, Sobrino F. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem. 1999;274:17580–17586. - PubMed
-
- Collinson LP, Dawes IW. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

