Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces
- PMID: 11410532
- PMCID: PMC312717
- DOI: 10.1101/gad.896201
Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces
Abstract
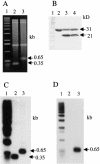
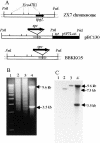
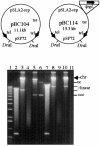
Linear plasmids and chromosomes of the bacterial genus Streptomyces have proteins of unknown characteristics and function linked covalently to their 5' DNA termini. We purified protein attached to the end of the pSLA2 linear plasmid of Streptomyces rochei, determined the N-terminal amino acid sequence, and used this information to clone corresponding genes from a S. rochei cosmid library. Three separate terminal protein genes (here designated as tpgR1, tpgR2, and tpgR3), which map to the S. rochei chromosome and to 100-kb and 206-kb linear plasmids contained in S. rochei, were isolated and found to encode a family of similar but distinct 21-kD proteins. Using tpgR1 to probe a genomic DNA library of Streptomyces lividans ZX7, whose linear chromosome can undergo transition to a circular form, we isolated a S. lividans chromosomal gene (tpgL) that we found specifies a protein closely related to, and functionally interchangeable with, TpgR proteins for pSLA2 maintenance in S. lividans. Mutation of tpgL precluded propagation of the pSLA2 plasmid in a linear form and also prevented propagation of S. lividans cells that contain linear, but not circular, chromosomes, indicating a specific and essential role for tpg genes in linear DNA replication. Surprisingly, Tpg proteins were observed to contain a reverse transcriptase-like domain rather than sequences in common with proteins that attach covalently to the termini of linear DNA replicons.
Figures


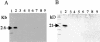
References
-
- Bibb MJ, Ward JM, Hopwood DA. Transformation of plasmid DNA into Streptomyces at high frequency. Nature. 1978;274:398–400. - PubMed
-
- Bibb MJ, Schottel JL, Cohen SN. A DNA cloning system for interspecies gene transfer in antibiotic-producing Streptomyces. Nature. 1980;284:526–531. - PubMed
-
- Bibb MJ, Findlay PR, Johnson MW. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. - PubMed
-
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
