Massive parallel analysis of the binding specificity of histone-like protein HU to single- and double-stranded DNA with generic oligodeoxyribonucleotide microchips
- PMID: 11410675
- PMCID: PMC55728
- DOI: 10.1093/nar/29.12.2654
Massive parallel analysis of the binding specificity of histone-like protein HU to single- and double-stranded DNA with generic oligodeoxyribonucleotide microchips
Abstract
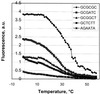
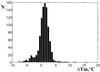
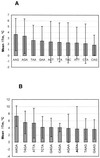
A generic hexadeoxyribonucleotide microchip has been applied to test the DNA-binding properties of HU histone-like bacterial protein, which is known to have a low sequence specificity. All 4096 hexamers flanked within 8mers by degenerate bases at both the 3'- and 5'-ends were immobilized within the 100 x 100 x 20 mm polyacrylamide gel pads of the microchip. Single-stranded immobilized oligonucleotides were converted in some experiments to the double-stranded form by hybridization with a specified mixture of 8mers. The DNA interaction with HU was characterized by three type of measurements: (i) binding of FITC-labeled HU to microchip oligonucleotides; (ii) melting curves of complexes of labeled HU with single-stranded microchip oligonucleotides; (iii) the effect of HU binding on melting curves of microchip double-stranded DNA labeled with another fluorescent dye, Texas Red. Large numbers of measurements of these parameters were carried out in parallel for all or many generic microchip elements in real time with a multi-wavelength fluorescence microscope. Statistical analysis of these data suggests some preference for HU binding to G/C-rich single-stranded oligonucleotides. HU complexes with double-stranded microchip 8mers can be divided into two groups in which HU binding either increased the melting temperature (T(m)) of duplexes or decreased it. The stabilized duplexes showed some preference for presence of the sequence motifs AAG, AGA and AAGA. In the second type of complex, enriched with A/T base pairs, the destabilization effect was higher for longer stretches of A/T duplexes. Binding of HU to labeled duplexes in the second type of complex caused some decrease in fluorescence. This decrease also correlates with the higher A/T content and lower T(m). The results demonstrate that generic microchips could be an efficient approach in analysis of sequence specificity of proteins.
Figures




Similar articles
-
Massive parallel analysis of DNA-Hoechst 33258 binding specificity with a generic oligodeoxyribonucleotide microchip.Nucleic Acids Res. 1999 Oct 15;27(20):4100-5. doi: 10.1093/nar/27.20.4100. Nucleic Acids Res. 1999. PMID: 10497276 Free PMC article.
-
Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips.Nucleic Acids Res. 1998 Mar 15;26(6):1515-21. doi: 10.1093/nar/26.6.1515. Nucleic Acids Res. 1998. PMID: 9490800 Free PMC article.
-
Specificity of mammalian Y-box binding protein p50 in interaction with ss and ds DNA analyzed with generic oligonucleotide microchip.J Mol Biol. 2002 Nov 15;324(1):73-87. doi: 10.1016/s0022-2836(02)00937-3. J Mol Biol. 2002. PMID: 12421560
-
[Biological microchips with hydrogel-immobilized nucleic acids, proteins, and other compounds: properties and applications in genomics].Mol Biol (Mosk). 2002 Jul-Aug;36(4):563-84. Mol Biol (Mosk). 2002. PMID: 12173458 Review. Russian.
-
Functional evolution of bacterial histone-like HU proteins.Curr Issues Mol Biol. 2011;13(1):1-12. Epub 2010 May 20. Curr Issues Mol Biol. 2011. PMID: 20484776 Review.
Cited by
-
Enhanced binding of an HU homologue under increased DNA supercoiling preserves chromosome organisation and sustains Streptomyces hyphal growth.Nucleic Acids Res. 2022 Nov 28;50(21):12202-12216. doi: 10.1093/nar/gkac1093. Nucleic Acids Res. 2022. PMID: 36420903 Free PMC article.
-
Quantitative prediction of NF-kappa B DNA-protein interactions.Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):8167-72. doi: 10.1073/pnas.102674699. Epub 2002 Jun 4. Proc Natl Acad Sci U S A. 2002. PMID: 12048232 Free PMC article.
-
A specific single-stranded DNA induces a distinct conformational change in the nucleoid-associated protein HU.Biochem Biophys Rep. 2016 Oct 11;8:318-324. doi: 10.1016/j.bbrep.2016.09.014. eCollection 2016 Dec. Biochem Biophys Rep. 2016. PMID: 28955971 Free PMC article.
-
Specificity assessment from fractionation experiments (SAFE): a novel method to evaluate microarray probe specificity based on hybridisation stringencies.Nucleic Acids Res. 2003 Jan 15;31(2):E1-1. doi: 10.1093/nar/gng001. Nucleic Acids Res. 2003. PMID: 12527790 Free PMC article.
-
Direct imaging of the circular chromosome in a live bacterium.Nat Commun. 2019 May 16;10(1):2194. doi: 10.1038/s41467-019-10221-0. Nat Commun. 2019. PMID: 31097704 Free PMC article.
References
-
- Maxam A., Mirzabekov,A. and Gilbert,W. (1976) Contact between the LAC repressor and DNA revealed by methylation. In Control of Ribosome Synthesis, Alfred Benzon Symposium IX. Munksgaard pp. 139–151.
-
- Fox K.R. (1997) DNase I fingerprinting. Methods Mol. Biol., 90, 1–22. - PubMed
-
- Petri V. and Brenowitz,M. (1997) Quantitative nucleic acids footprinting thermodynamic and kinetic approaches. Curr. Opin. Biotechnol., 8, 36–44. - PubMed
-
- Zasedatelev A.S., Mikhailov,M.V., Krylov,A.S. and Gursky,G.V. (1980) Mechanism of recognition of AT-pairs in DNA molecules of the dye Hoechst 33258. Dokl. Akad. Nauk SSSR, 255, 756–760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

