Viral replicase gene products suffice for coronavirus discontinuous transcription
- PMID: 11413334
- PMCID: PMC114390
- DOI: 10.1128/JVI.75.14.6676-6681.2001
Viral replicase gene products suffice for coronavirus discontinuous transcription
Abstract
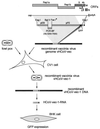
We have used vaccinia virus as a vector to clone a 22.5-kbp cDNA that represents the 5' and 3' ends of the human coronavirus 229E (HCoV 229E) genome, the HCoV 229E replicase gene, and a single reporter gene (coding for green fluorescent protein [GFP]) located downstream of a regulatory element for coronavirus mRNA transcription. When RNA transcribed from this cDNA was transfected into BHK-21 cells, a small percentage of cells displayed strong fluorescence. A region of the mRNA encoding GFP was amplified by PCR and shown to have the unique mRNA leader-body junction indicative of coronavirus-mediated transcription. These data show that the coronavirus replicase gene products suffice for discontinuous subgenomic mRNA transcription.
Figures


References
-
- Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

