Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control
- PMID: 11416129
- PMCID: PMC87109
- DOI: 10.1128/MCB.21.14.4495-4504.2001
Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control
Abstract
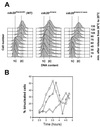
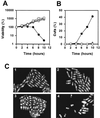
In Schizosaccharomyces pombe, the catalytic subunit of DNA polymerase epsilon (Pol epsilon) is encoded by cdc20(+) and is essential for chromosomal DNA replication. Here we demonstrate that the N-terminal half of Pol epsilon that includes the highly conserved polymerase and exonuclease domains is dispensable for cell viability, similar to observations made with regard to Saccharomyces cerevisiae. However, unlike budding yeast, we find that fission yeast cells lacking the N terminus of Pol epsilon (cdc20(DeltaN-term)) are hypersensitive to DNA-damaging agents and have a cell cycle delay. Moreover, the viability of cdc20(DeltaN-term) cells is dependent on expression of rad3(+), hus1(+), and chk1(+), three genes essential for the DNA damage checkpoint control. These data suggest that in the absence of the N terminus of Pol epsilon, cells accumulate DNA damage that must be repaired prior to mitosis. Our observation that S phase occurs more slowly for cdc20(DeltaN-term) cells suggests that DNA damage might result from defects in DNA synthesis. We hypothesize that the C-terminal half of Pol epsilon is required for assembly of the replicative complex at the onset of S phase. This unique and essential function of the C terminus is preserved in the absence of the N-terminal catalytic domains, suggesting that the C terminus can interact with and recruit other DNA polymerases to the site of initiation.
Figures



References
-
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
