Evolution of gene order conservation in prokaryotes
- PMID: 11423009
- PMCID: PMC33396
- DOI: 10.1186/gb-2001-2-6-research0020
Evolution of gene order conservation in prokaryotes
Abstract
Background: As more complete genomes are sequenced, conservation of gene order between different organisms is emerging as an informative property of the genomes. Conservation of gene order has been used for predicting function and functional interactions of proteins, as well as for studying the evolutionary relationships between genomes. The reasons for the maintenance of gene order are still not well understood, as the organization of the prokaryote genome into operons and lateral gene transfer cannot possibly account for all the instances of conservation found. Comprehensive studies of gene order are one way of elucidating the nature of these maintaining forces.
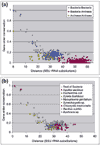
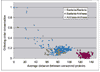
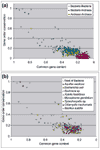
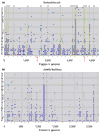
Results: Gene order is extensively conserved between closely related species, but rapidly becomes less conserved among more distantly related organisms, probably in a cooperative fashion. This trend could be universal in prokaryotic genomes, as archaeal genomes are likely to behave similarly to bacterial genomes. Gene order conservation could therefore be used as a valid phylogenetic measure to study relationships between species. Even between very distant species, remnants of gene order conservation exist in the form of highly conserved clusters of genes. This suggests the existence of selective processes that maintain the organization of these regions. Because the clusters often span more than one operon, common regulation probably cannot be invoked as the cause of the maintenance of gene order.
Conclusions: Gene order conservation is a genomic measure that can be useful for studying relationships between prokaryotes and the evolutionary forces shaping their genomes. Gene organization is extensively conserved in some genomic regions, and further studies are needed to elucidate the reason for this conservation.
Figures

Similar articles
-
Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context.Genome Res. 2001 Mar;11(3):356-72. doi: 10.1101/gr.gr-1619r. Genome Res. 2001. PMID: 11230160
-
Inferring Functional Relationships from Conservation of Gene Order.Methods Mol Biol. 2017;1526:41-63. doi: 10.1007/978-1-4939-6613-4_3. Methods Mol Biol. 2017. PMID: 27896735
-
Inferring functional relationships from conservation of gene order.Methods Mol Biol. 2008;453:181-99. doi: 10.1007/978-1-60327-429-6_8. Methods Mol Biol. 2008. PMID: 18712303
-
Ancient origin of the tryptophan operon and the dynamics of evolutionary change.Microbiol Mol Biol Rev. 2003 Sep;67(3):303-42, table of contents. doi: 10.1128/MMBR.67.3.303-342.2003. Microbiol Mol Biol Rev. 2003. PMID: 12966138 Free PMC article. Review.
-
Prokaryotic genomes: the emerging paradigm of genome-based microbiology.Curr Opin Genet Dev. 1997 Dec;7(6):757-63. doi: 10.1016/s0959-437x(97)80037-8. Curr Opin Genet Dev. 1997. PMID: 9468784 Review.
Cited by
-
G-NEST: a gene neighborhood scoring tool to identify co-conserved, co-expressed genes.BMC Bioinformatics. 2012 Sep 28;13:253. doi: 10.1186/1471-2105-13-253. BMC Bioinformatics. 2012. PMID: 23020263 Free PMC article.
-
The complete genome sequence of the pathogenic intestinal spirochete Brachyspira pilosicoli and comparison with other Brachyspira genomes.PLoS One. 2010 Jul 6;5(7):e11455. doi: 10.1371/journal.pone.0011455. PLoS One. 2010. PMID: 20625514 Free PMC article.
-
argC Orthologs from Rhizobiales show diverse profiles of transcriptional efficiency and functionality in Sinorhizobium meliloti.J Bacteriol. 2011 Jan;193(2):460-72. doi: 10.1128/JB.01010-10. Epub 2010 Nov 12. J Bacteriol. 2011. PMID: 21075924 Free PMC article.
-
Uncovering major genomic features of essential genes in Bacteria and a methanogenic Archaea.FEBS J. 2015 Sep;282(17):3395-3411. doi: 10.1111/febs.13350. Epub 2015 Jul 14. FEBS J. 2015. PMID: 26084810 Free PMC article.
-
Toward the Complete Functional Characterization of a Minimal Bacterial Proteome.J Phys Chem B. 2022 Sep 15;126(36):6820-6834. doi: 10.1021/acs.jpcb.2c04188. Epub 2022 Sep 1. J Phys Chem B. 2022. PMID: 36048731 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

