Antifungal proteins
- PMID: 11425698
- PMCID: PMC92957
- DOI: 10.1128/AEM.67.7.2883-2894.2001
Antifungal proteins
Figures
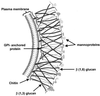




References
-
- Agrawal G K, Jwa N S, Rakwal R. A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem Biophys Res Commun. 2000;274:157–165. - PubMed
-
- Ahmed A, Sesti F, Ilan N, Shih T M, Sturley S L, Goldstein S A. A molecular target for viral killer toxin: TOK1 potassium channels. Cell. 1999;99:283–291. - PubMed
-
- Ainsworth G, editor. The fungi, an advance treatise. New York, N.Y: Academic Press; 1973.
-
- Almeida M S, Cabral K M, Zingali R B, Kurtenbach E. Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch Biochem Biophys. 2000;378:278–286. - PubMed
-
- Alves A L, De Samblanx G W, Terras R R, Cammue B P, Broekaert W F. Expression of functional Raphanus sativusantifungal protein in yeast. FEBS Lett. 1994;348:228–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

