Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments
- PMID: 11425703
- PMCID: PMC92962
- DOI: 10.1128/AEM.67.7.2922-2926.2001
Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments
Abstract
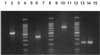
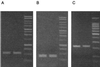
Primer sets were designed to target specific 16S ribosomal DNA (rDNA) sequences of photosynthetic bacteria, including the green sulfur bacteria, the green nonsulfur bacteria, and the members of the Heliobacteriaceae (a gram-positive phylum). Due to the phylogenetic diversity of purple sulfur and purple nonsulfur phototrophs, the 16S rDNA gene was not an appropriate target for phylogenetic rDNA primers. Thus, a primer set was designed that targets the pufM gene, encoding the M subunit of the photosynthetic reaction center, which is universally distributed among purple phototrophic bacteria. The pufM primer set amplified DNAs not only from purple sulfur and purple nonsulfur phototrophs but also from Chloroflexus species, which also produce a reaction center like that of the purple bacteria. Although the purple bacterial reaction center structurally resembles green plant photosystem II, the pufM primers did not amplify cyanobacterial DNA, further indicating their specificity for purple anoxyphototrophs. This combination of phylogenetic- and photosynthesis-specific primers covers all groups of known anoxygenic phototrophs and as such shows promise as a molecular tool for the rapid assessment of natural samples in ecological studies of these organisms.
Figures

References
-
- Bryantseva I A, Gorlenko V M, Kompantseva E I, Achenbach L A, Madigan M T. Heliorestis daurensis gen. nov. sp. nov., an alkaliphilic coiled to rod-shaped phototrophic heliobacterium from an alkaline Siberian soda lake. Arch Microbiol. 1999;172:167–174. - PubMed
-
- Casamayor E O, Calderón-Paz J I, Mas J, Pedrós-Alió C. Identification of phototrophic sulfur bacteria through the analysis of lmwRNA band patterns. Arch Microbiol. 1998;170:269–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

