Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis
- PMID: 11425708
- PMCID: PMC92967
- DOI: 10.1128/AEM.67.7.2958-2965.2001
Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis
Abstract
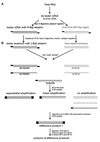
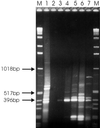
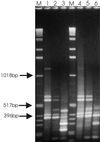
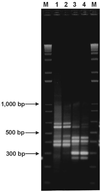
Microbial proliferation and biofilm formation on biologic or inert substrates are characteristics of invasive Staphylococcus aureus infections and is associated with phenotypic alterations such as reduced antimicrobial susceptibility. To identify genes which are typically expressed in biofilms, a micro-representational-difference analysis (micro-RDA) was adapted for gram-positive bacteria and used with cDNA derived from populations of S. aureus DSM 20231 growing in a biofilm or plankonically. In comparison to previously described cDNA RDA protocols, micro-RDA has the advantages that only minimal quantities of total RNA are needed and, most importantly, that total RNA can be used since the large amount of rRNA in total RNA does not interfere with the micro-RDA procedure. Using a series of spiked controls with various amounts of MS2 RNA in a background of total RNA from S. aureus, the equivalent of five copies of MS2 per cell were detectable after three rounds of subtractive enrichment. Five genes were identified as being differentially expressed in biofilm versus planktonic cultures. These genes revealed homology to a threonyl-tRNA synthetase, a phosphoglycerate mutase, a triosephosphate isomerase, an alcohol dehydrogenase I, and a ClpC ATPase. Differential levels of expression were subsequently confirmed by standard Northern blotting. In conclusion, micro-RDA is a sensitive and specific method to detect transcripts differentially expressed as a function of different S. aureus growth conditions.
Figures

References
-
- Bowler L D, Hubank M, Spratt B G. Representational difference analysis of cDNA for the detection of differential gene expression in bacteria: development using a model of iron-regulated gene expression in Neisseria meningitidis. Microbiology. 1999;145:3529–3537. - PubMed
-
- Brown M R, Allison D G, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. - PubMed
-
- Brown M R, Williams P. Influence of substrate limitation and growth phase on sensitivity to antimicrobial agents. J Antimicrob Chemother. 1985;15(Suppl. A):7–14. - PubMed
-
- Charpentier E, Novak R, Tuomanen E. Regulation of growth inhibition at high temperature, autolysis, transformation and adherence in Streptococcus pneumoniae by clpC. Mol Microbiol. 2000;37:717–726. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

