Microarray analysis of microbial virulence factors
- PMID: 11425749
- PMCID: PMC93008
- DOI: 10.1128/AEM.67.7.3258-3263.2001
Microarray analysis of microbial virulence factors
Abstract
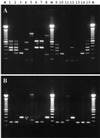
Hybridization with oligonucleotide microchips (microarrays) was used for discrimination among strains of Escherichia coli and other pathogenic enteric bacteria harboring various virulence factors. Oligonucleotide microchips are miniature arrays of gene-specific oligonucleotide probes immobilized on a glass surface. The combination of this technique with the amplification of genetic material by PCR is a powerful tool for the detection of and simultaneous discrimination among food-borne human pathogens. The presence of six genes (eaeA, slt-I, slt-II, fliC, rfbE, and ipaH) encoding bacterial antigenic determinants and virulence factors of bacterial strains was monitored by multiplex PCR followed by hybridization of the denatured PCR product to the gene-specific oligonucleotides on the microchip. The assay was able to detect these virulence factors in 15 Salmonella, Shigella, and E. coli strains. The results of the chip analysis were confirmed by hybridization of radiolabeled gene-specific probes to genomic DNA from bacterial colonies. In contrast, gel electrophoretic analysis of the multiplex PCR products used for the microarray analysis produced ambiguous results due to the presence of unexpected and uncharacterized bands. Our results suggest that microarray analysis of microbial virulence factors might be very useful for automated identification and characterization of bacterial pathogens.
Figures


References
-
- Cebula T A, Koch W H. Analysis of spontaneous and psoralen-induced Salmonella typhimurium hisG46 revertants by oligodeoxyribonucleotide colony hybridization: use of psoralens to cross-link probes to target sequences. Mutat Res. 1990;229:79–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

