Differential expression of genes encoding subthreshold-operating voltage-gated K+ channels in brain
- PMID: 11425889
- PMCID: PMC6762370
- DOI: 10.1523/JNEUROSCI.21-13-04609.2001
Differential expression of genes encoding subthreshold-operating voltage-gated K+ channels in brain
Abstract
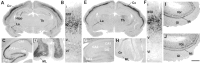
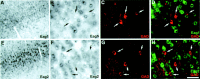
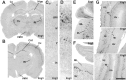
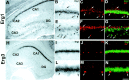
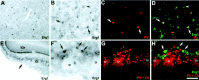
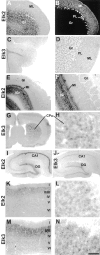
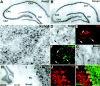
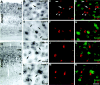
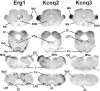
The members of the three subfamilies (eag, erg, and elk) of the ether-a-go-go (EAG) family of potassium channel pore-forming subunits express currents that, like the M-current (I(M)), could have considerable influence on the subthreshold properties of neuronal membranes, and hence the control of excitability. A nonradioactive in situ hybridization (NR-ISH) study of the distribution of the transcripts encoding the eight known EAG family subunits in rat brain was performed to identify neuronal populations in which the physiological roles of EAG channels could be studied. These distributions were compared with those of the mRNAs encoding the components of the classical M-current (Kcnq2 and Kcnq3). NR-ISH was combined with immunohistochemistry to specific neuronal markers to help identify expressing neurons. The results show that each EAG subunit has a specific pattern of expression in rat brain. EAG and Kcnq transcripts are prominent in several types of excitatory neurons in the cortex and hippocampus; however, only one of these channel components (erg1) was consistently expressed in inhibitory interneurons in these areas. Some neuronal populations express more than one product of the same subfamily, suggesting that the subunits may form heteromeric channels in these neurons. Many neurons expressed multiple EAG family and Kcnq transcripts, such as CA1 pyramidal neurons, which contained Kcnq2, Kcnq3, eag1, erg1, erg3, elk2, and elk3. This indicates that the subthreshold current in many neurons may be complex, containing different components mediated by a number of channels with distinct properties and neuromodulatory responses.
Figures



References
-
- Abbott GW, Sesti F, Splawski I, Buck ME, Lehmann MH, Timothy KW, Keating MT, Goldstein SA. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97:175–187. - PubMed
-
- Baxter DA, Byrne JH. Ionic conductance mechanisms contributing to the electrophysiological properties of neurons. Curr Opin Neurobiol. 1991;1:105–112. - PubMed
-
- Brown DA. M channels. In: Narahashi ET, editor. Ion channels. Plenum; New York: 1988. pp. 55–94. - PubMed
-
- Brown DA, Adams PR. Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature. 1980;283:673–676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
