The OMP-lacZ transgene mimics the unusual expression pattern of OR-Z6, a new odorant receptor gene on mouse chromosome 6: implication for locus-dependent gene expression
- PMID: 11425891
- PMCID: PMC6762339
- DOI: 10.1523/JNEUROSCI.21-13-04637.2001
The OMP-lacZ transgene mimics the unusual expression pattern of OR-Z6, a new odorant receptor gene on mouse chromosome 6: implication for locus-dependent gene expression
Erratum in
- J Neurosci 2001 Aug 15;21(16):6457
Abstract
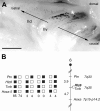
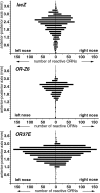
Reporter gene expression in the olfactory epithelium of H-lacZ6 transgenic mice mimics the cell-selective expression pattern known for some odorant receptor genes. The transgene construct in these mice consists of the lacZ coding region, driven by the proximal olfactory marker protein (OMP) gene promoter, and shows expression in a zonally confined subpopulation of olfactory neurons. To address mechanisms underlying the odorant receptor-like expression pattern of the lacZ construct, we analyzed the transgene-flanking region and identified OR-Z6, the first cloned odorant receptor gene that maps to mouse chromosome 6. OR-Z6 bears the highest sequence similarity (85%) to a human odorant receptor gene at the syntenic location on human chromosome 7. We analyzed the expression pattern of OR-Z6 in olfactory tissues of H-lacZ6 mice and show that it bears strong similarities to that mapped for beta-galactosidase. Expression of both genes in olfactory neurons is primarily restricted to the same medial subregion of the olfactory epithelium. Axons from both neuronal subpopulations project to the same ventromedial aspect of the anterior olfactory bulbs. Furthermore, colocalization analyses in H-lacZ6 mice demonstrate that OR-Z6-reactive glomeruli receive axonal input from lacZ-positive neurons as well. These results suggest that the expression of both genes is coordinated and that transgene expression in H-lacZ6 mice is regulated by locus-dependent mechanisms.
Figures






References
-
- Asai H, Kasai H, Matsuda Y, Yamazaki N, Nagawa F, Sakano H, Tsuboi A. Genomic structure and transcription of a murine odorant receptor gene: differential initiation of transcription in the olfactory and testicular cells. Biochem Biophys Res Commun. 1996;221:240–247. - PubMed
-
- Astic L, Saucier D. Anatomical mapping of the neuroepithelial projection to the olfactory bulb in the rat. Brain Res Bull. 1986;16:445–454. - PubMed
-
- Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter DH, Carrozzo R, Patel K, Sheer D, Lehrach H. Olfactory receptor gene cluster on human chromosome 17: possible duplication of an ancestral receptor repertoire. Hum Mol Genet. 1994;3:229–235. - PubMed
-
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a basis for odorant recognition. Cell. 1991;65:175–187. - PubMed
-
- Carver EA, Issel-Tarver L, Rine J, Olsen AS, Stubbs L. Location of mouse and human genes corresponding to conserved canine olfactory receptor gene family. Mamm Genome. 1998;9:349–354. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
