rpoB sequence analysis of cultured Tropheryma whippelii
- PMID: 11427549
- PMCID: PMC88165
- DOI: 10.1128/JCM.39.7.2425-2430.2001
rpoB sequence analysis of cultured Tropheryma whippelii
Abstract
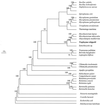
Until recently no isolate of Tropheryma whippelii was available, and therefore genetic studies were limited to those based on PCR amplification of conserved genes. In this study we determined the nucleotide sequence of rpoB (encoding the beta-subunit of RNA polymerase) from a cultured strain of T. whippelii using degenerate consensus PCR and genome walking. The T. whippelii rpoB consists of 3,657 bp with a 50.4% GC content and encodes 1,218 amino acids with a calculated molecular mass of 138 kDa. Comparison of T. whippelii RpoB with other eubacterial RpoB proteins indicated sequence similarity ranging from 57.19 (Mycoplasma pneumoniae) to 74.63% (Mycobacterium tuberculosis). Phylogenetic analysis of T. whippelii based on comparison of its RpoB sequence with sequences available for other bacteria was consistent with that previously derived from the 16S ribosomal DNA (rDNA) sequence, indicating that it belongs to the actinomyces clade. The sequence comparison allowed the design of a primer pair, TwrpoB.F and TwrpoB.R, specific for T. whippelii rpoB. When incorporated into a PCR, this primer pair allowed the detection of T. whippelii rpoB in three of three 16S rDNA PCR-positive biopsy specimens and zero of seven negative controls. rpoB could therefore be targeted in PCR-mediated detection and identification of this emerging bacterial species. This approach has previously been shown useful for the identification of related mycobacteria. This study underscores that a method involving isolation and then propagation of emerging bacteria is a useful way to quickly achieve extensive molecular knowledge of these pathogens.
Figures



References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

