The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia
- PMID: 11427734
- PMCID: PMC35443
- DOI: 10.1073/pnas.141234698
The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia
Abstract
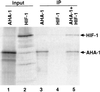
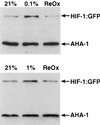
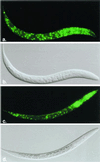
Hypoxia-inducible factor, a heterodimeric transcription complex, regulates cellular and systemic responses to low oxygen levels (hypoxia) during normal mammalian development or tumor progression. Here, we present evidence that a similar complex mediates response to hypoxia in Caenorhabditis elegans. This complex consists of HIF-1 and AHA-1, which are encoded by C. elegans homologs of the hypoxia-inducible factor (HIF) alpha and beta subunits, respectively. hif-1 mutants exhibit no severe defects under standard laboratory conditions, but they are unable to adapt to hypoxia. Although wild-type animals can survive and reproduce in 1% oxygen, the majority of hif-1-defective animals die in these conditions. We show that the expression of an HIF-1:green fluorescent protein fusion protein is induced by hypoxia and is subsequently reduced upon reoxygenation. Both hif-1 and aha-1 are expressed in most cell types, and the gene products can be coimmunoprecipitated. We conclude that the mechanisms of hypoxia signaling are likely conserved among metazoans. Additionally, we find that nuclear localization of AHA-1 is disrupted in an hif-1 mutant. This finding suggests that heterodimerization may be a prerequisite for efficient nuclear translocation of AHA-1.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

