The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond
- PMID: 11431476
- PMCID: PMC8009867
- DOI: 10.1074/jbc.M104097200
The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond
Abstract
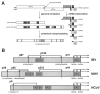
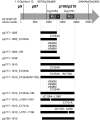
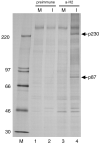
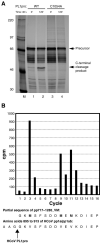
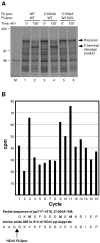
The largest replicative protein of coronaviruses is known as p195 in the avian infectious bronchitis virus (IBV) and p210 (p240) in the mouse hepatitis virus. It is autocatalytically released from the precursors pp1a and pp1ab by one zinc finger-containing papain-like protease (PLpro) in IBV and by two paralogous PLpros, PL1pro and PL2pro, in mouse hepatitis virus. The PLpro-containing proteins have been recently implicated in the control of coronavirus subgenomic mRNA synthesis (transcription). By using comparative sequence analysis, we now show that the respective proteins of all sequenced coronaviruses are flanked by two conserved PLpro cleavage sites and share a complex (multi)domain organization with PL1pro being inactivated in IBV. Based upon these predictions, the processing of the human coronavirus 229E p195/p210 N terminus was studied in detail. First, an 87-kDa protein (p87), which is derived from a pp1a/pp1ab region immediately upstream of p195/p210, was identified in human coronavirus 229E-infected cells. Second, in vitro synthesized proteins representing different parts of pp1a were autocatalytically processed at the predicted site. Surprisingly, both PL1pro and PL2pro cleaved between p87 and p195/p210. The PL1pro-mediated cleavage was slow and significantly suppressed by a non-proteolytic activity of PL2pro. In contrast, PL2pro, whose proteolytic activity and specificity were established in this study, cleaved the same site efficiently in the presence of the upstream domains. Third, a correlation was observed between the overlapping substrate specificities and the parallel evolution of PL1pro and PL2pro. Collectively, our results imply that the p195/p210 autoprocessing mechanisms may be conserved among coronaviruses to an extent not appreciated previously, with PL2pro playing a major role. A large subset of coronaviruses may employ two proteases to cleave the same site(s) and thus regulate the expression of the viral genome in a unique way.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

