Biology of Giardia lamblia
- PMID: 11432808
- PMCID: PMC88984
- DOI: 10.1128/CMR.14.3.447-475.2001
Biology of Giardia lamblia
Abstract
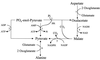
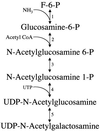
Giardia lamblia is a common cause of diarrhea in humans and other mammals throughout the world. It can be distinguished from other Giardia species by light or electron microscopy. The two major genotypes of G. lamblia that infect humans are so different genetically and biologically that they may warrant separate species or subspecies designations. Trophozoites have nuclei and a well-developed cytoskeleton but lack mitochondria, peroxisomes, and the components of oxidative phosphorylation. They have an endomembrane system with at least some characteristics of the Golgi complex and encoplasmic reticulum, which becomes more extensive in encysting organisms. The primitive nature of the organelles and metabolism, as well as small-subunit rRNA phylogeny, has led to the proposal that Giardia spp. are among the most primitive eukaryotes. G. lamblia probably has a ploidy of 4 and a genome size of approximately 10 to 12 Mb divided among five chromosomes. Most genes have short 5' and 3' untranslated regions and promoter regions that are near the initiation codon. Trophozoites exhibit antigenic variation of an extensive repertoire of cysteine-rich variant-specific surface proteins. Expression is allele specific, and changes in expression from one vsp gene to another have not been associated with sequence alterations or gene rearrangements. The Giardia genome project promises to greatly increase our understanding of this interesting and enigmatic organism.
Figures





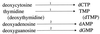





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

