Listeria pathogenesis and molecular virulence determinants
- PMID: 11432815
- PMCID: PMC88991
- DOI: 10.1128/CMR.14.3.584-640.2001
Listeria pathogenesis and molecular virulence determinants
Abstract
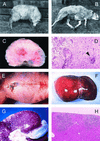
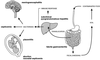
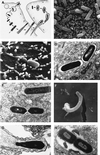
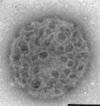
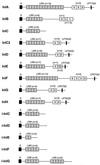
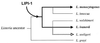
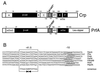
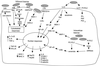
The gram-positive bacterium Listeria monocytogenes is the causative agent of listeriosis, a highly fatal opportunistic foodborne infection. Pregnant women, neonates, the elderly, and debilitated or immunocompromised patients in general are predominantly affected, although the disease can also develop in normal individuals. Clinical manifestations of invasive listeriosis are usually severe and include abortion, sepsis, and meningoencephalitis. Listeriosis can also manifest as a febrile gastroenteritis syndrome. In addition to humans, L. monocytogenes affects many vertebrate species, including birds. Listeria ivanovii, a second pathogenic species of the genus, is specific for ruminants. Our current view of the pathophysiology of listeriosis derives largely from studies with the mouse infection model. Pathogenic listeriae enter the host primarily through the intestine. The liver is thought to be their first target organ after intestinal translocation. In the liver, listeriae actively multiply until the infection is controlled by a cell-mediated immune response. This initial, subclinical step of listeriosis is thought to be common due to the frequent presence of pathogenic L. monocytogenes in food. In normal individuals, the continual exposure to listerial antigens probably contributes to the maintenance of anti-Listeria memory T cells. However, in debilitated and immunocompromised patients, the unrestricted proliferation of listeriae in the liver may result in prolonged low-level bacteremia, leading to invasion of the preferred secondary target organs (the brain and the gravid uterus) and to overt clinical disease. L. monocytogenes and L. ivanovii are facultative intracellular parasites able to survive in macrophages and to invade a variety of normally nonphagocytic cells, such as epithelial cells, hepatocytes, and endothelial cells. In all these cell types, pathogenic listeriae go through an intracellular life cycle involving early escape from the phagocytic vacuole, rapid intracytoplasmic multiplication, bacterially induced actin-based motility, and direct spread to neighboring cells, in which they reinitiate the cycle. In this way, listeriae disseminate in host tissues sheltered from the humoral arm of the immune system. Over the last 15 years, a number of virulence factors involved in key steps of this intracellular life cycle have been identified. This review describes in detail the molecular determinants of Listeria virulence and their mechanism of action and summarizes the current knowledge on the pathophysiology of listeriosis and the cell biology and host cell responses to Listeria infection. This article provides an updated perspective of the development of our understanding of Listeria pathogenesis from the first molecular genetic analyses of virulence mechanisms reported in 1985 until the start of the genomic era of Listeria research.
Figures

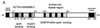
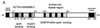




References
-
- Abram M, Doric M. Primary Listeria monocytogenes infection in gestating mice. Folia Microbiol. 1997;42:65–71. - PubMed
-
- Agranof D D, Krishna S. Metal ion homeostasis and intracellular parasitism. Mol Microbiol. 1998;28:403–412. - PubMed
-
- Akaki T, Sato K, Shimizu T, Sano C, Kajitani H, Dekio S, Tomioka H. Effector molecules in expression of the antimicrobial activity of macrophages against Mycobacterium avium complex: roles of reactive nitrogen intermediates, reactive oxygen intermediates, and free fatty acids. J Leukocyte Biol. 1997;62:795–804. - PubMed
-
- Alexander A V, Walker R L, Johnson B J, Charlton B R, Woods L W. Bovine abortions attributable to Listeria ivanovii: four cases (1988–1990) J Am Vet Med Assoc. 1992;200:711–714. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

