Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN
- PMID: 11432837
- PMCID: PMC125512
- DOI: 10.1093/emboj/20.13.3506
Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN
Abstract
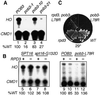
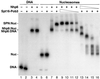
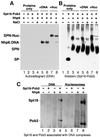
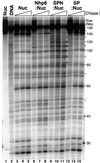
Yeast Spt16/Cdc68 and Pob3 form a heterodimer that acts in both DNA replication and transcription. This is supported by studies of new alleles of SPT16 described here. We show that Spt16-Pob3 enhances HO transcription through a mechanism that is affected by chromatin modification, since some of the defects caused by mutations can be suppressed by deleting the histone deacetylase Rpd3. While otherwise conserved among many eukaryotes, Pob3 lacks the HMG1 DNA-binding motif found in similar proteins such as the SSRP1 subunit of human FACT. SPT16 and POB3 display strong genetic interactions with NHP6A/B, which encodes an HMG1 motif, suggesting that these gene products function coordinately in vivo. While Spt16-Pob3 and Nhp6 do not appear to form stable heterotrimers, Nhp6 binds to nucleosomes and these Nhp6-nucleosomes can recruit Spt16-Pob3 to form SPN-nucleosomes. These complexes have altered electrophoretic mobility and a distinct pattern of enhanced sensitivity to DNase I. These results suggest that Spt16-Pob3 and Nhp6 cooperate to function as a novel nucleosome reorganizing factor.
Figures


References
-
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1994) Current Protocols in Molecular Biology. Greene Publishing Associates, Inc., and John Wiley & Sons, Inc., Boston, MA.
-
- Bartel P.L. and Fields,S. (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol., 254, 241–263. - PubMed
-
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. - PubMed
-
- Brazas R.M., Bhoite,L.T., Murphy,M.D., Yu,Y., Chen,Y., Neklason,D.W. and Stillman,D.J. (1995) Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J. Biol. Chem., 270, 29151–29161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

