An upstream repressor element plays a role in Igf2 imprinting
- PMID: 11432838
- PMCID: PMC125515
- DOI: 10.1093/emboj/20.13.3518
An upstream repressor element plays a role in Igf2 imprinting
Abstract
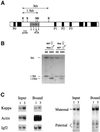
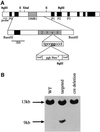
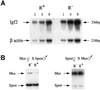
The imprinted Igf2 gene is associated with a small upstream region that is differentially methylated on the active paternal allele. We have identified a repressor element within this sequence and shown that repression is probably mediated through a trans- acting factor, GCF2. DNA methylation of this site abrogates both protein binding and repressor activity. Targeting experiments demonstrate that this element plays a role in the repression of the maternal Igf2 gene in vivo.
Figures

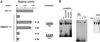
Similar articles
-
Epigenetic regulation of Igf2/H19 imprinting at CTCF insulator binding sites.J Cell Biochem. 2003 Dec 1;90(5):1038-55. doi: 10.1002/jcb.10684. J Cell Biochem. 2003. PMID: 14624463
-
Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene.Nature. 2000 May 25;405(6785):482-5. doi: 10.1038/35013100. Nature. 2000. PMID: 10839546
-
CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus.Hum Mol Genet. 2006 Oct 1;15(19):2945-54. doi: 10.1093/hmg/ddl237. Epub 2006 Aug 23. Hum Mol Genet. 2006. PMID: 16928784
-
IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch.Mol Hum Reprod. 2005 Sep;11(9):631-40. doi: 10.1093/molehr/gah230. Epub 2005 Oct 11. Mol Hum Reprod. 2005. PMID: 16219628
-
Chromatin structure and imprinting: developmental control of DNase-I sensitivity in the mouse insulin-like growth factor 2 gene.Dev Genet. 1995;17(3):240-52. doi: 10.1002/dvg.1020170309. Dev Genet. 1995. PMID: 8565330
Cited by
-
DNA methylation and its role in the pathogenesis of diabetes.Pediatr Diabetes. 2017 May;18(3):167-177. doi: 10.1111/pedi.12521. Pediatr Diabetes. 2017. PMID: 28401680 Free PMC article. Review.
-
Callipyge mutation affects gene expression in cis: a potential role for chromatin structure.Genome Res. 2006 Mar;16(3):340-6. doi: 10.1101/gr.4389306. Epub 2006 Jan 13. Genome Res. 2006. PMID: 16415109 Free PMC article.
-
DNA methylation may restrict but does not determine differential gene expression at the Sgy/Tead2 locus during mouse development.Mol Cell Biol. 2004 Mar;24(5):1968-82. doi: 10.1128/MCB.24.5.1968-1982.2004. Mol Cell Biol. 2004. PMID: 14966277 Free PMC article.
-
Structural and functional analysis of a 0.5-Mb chicken region orthologous to the imprinted mammalian Ascl2/Mash2-Igf2-H19 region.Genome Res. 2005 Jan;15(1):154-65. doi: 10.1101/gr.2609605. Epub 2004 Dec 8. Genome Res. 2005. PMID: 15590938 Free PMC article.
-
Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development.J Biol Chem. 2014 Aug 22;289(34):23882-92. doi: 10.1074/jbc.M114.573469. Epub 2014 Jul 11. J Biol Chem. 2014. PMID: 25016019 Free PMC article.
References
-
- Bartolomei M.S., Webber,A.L., Brunkow,M.E. and Tilghman,S.M. (1993) Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev., 7, 1663–1673. - PubMed
-
- Borras T., Peterson,C.A. and Piatigorsky,J. (1988) Evidence for positive and negative regulation in the promoter of the chicken δ1-crystallin gene. Dev. Biol., 127, 209–219. - PubMed
-
- Constancia M., Pickard,B., Kelsey,G. and Reik,W. (1998) Imprinting mechanisms. Genome Res., 8, 881–900. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

