Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex
- PMID: 11433020
- PMCID: PMC55777
- DOI: 10.1093/nar/29.13.2757
Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex
Abstract
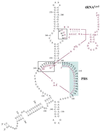
Human immunodeficiency virus type 2 (HIV-2) reverse transcription is initiated from cellular tRNA(Lys3) partially annealed to the RNA viral genome at the primer binding site (PBS). This annealing involves interactions between two highly structured RNA molecules. In contrast to HIV-1, in which the reverse transcription initiation complex has been thoroughly studied, there is still little information regarding a possible model to describe the secondary structure of the template-primer complex in HIV-2. To determine whether HIV-2 RNA sequences flanking the PBS may specifically interact with the natural primer tRNA, we performed site-directed mutagenesis and enzymatic footprinting. An RNA fragment corresponding to the HIV-2 U5 RNA domain and tRNA(Lys3) were probed either in their free form or in the binary complex. Important reactivity changes to nucleases were obtained upon complex formation. In addition to the canonical contacts between the viral PBS and the 3' end acceptor stem of tRNA(Lys3), we identified two additional interacting domains: (i) the U-rich region of the anticodon loop with the A-rich sequence of the internal loop within the U5-prePBS region; (ii) nucleotides 48-54 from the TPsiC domain of tRNA(Lys3) and the 240-247 region of viral U5-RNA. In view of these experimental data and sequence comparison between different HIV-2 isolates, we propose a model for the secondary structure of the HIV-2 template-primer initiation complex.
Figures









Similar articles
-
A structured RNA motif is involved in correct placement of the tRNA(3)(Lys) primer onto the human immunodeficiency virus genome.J Virol. 2000 Mar;74(5):2227-38. doi: 10.1128/jvi.74.5.2227-2238.2000. J Virol. 2000. PMID: 10666253 Free PMC article.
-
tRNA(Lys3): the primer tRNA for reverse transcription in HIV-1.IUBMB Life. 2002 Feb;53(2):107-14. doi: 10.1080/15216540211469. IUBMB Life. 2002. PMID: 12049192 Review.
-
Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS.RNA. 2002 Mar;8(3):357-69. doi: 10.1017/s1355838202028194. RNA. 2002. PMID: 12003495 Free PMC article.
-
Human immunodeficiency virus Type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA(Lys3) in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site.J Virol. 1996 Aug;70(8):4996-5004. doi: 10.1128/JVI.70.8.4996-5004.1996. J Virol. 1996. PMID: 8764006 Free PMC article.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
Cited by
-
Synthetic tRNALys,3 as the replication primer for the HIV-1HXB2 and HIV-1Mal genomes.Nucleic Acids Res. 2004 Sep 1;32(15):4687-95. doi: 10.1093/nar/gkh813. Print 2004. Nucleic Acids Res. 2004. PMID: 15342789 Free PMC article.
-
Structure-function relationships of the initiation complex of HIV-1 reverse transcription: the case of mutant viruses using tRNA(His) as primer.Nucleic Acids Res. 2003 Oct 1;31(19):5764-75. doi: 10.1093/nar/gkg754. Nucleic Acids Res. 2003. PMID: 14500840 Free PMC article.
-
Forced selection of a human immunodeficiency virus type 1 variant that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme.J Virol. 2004 Oct;78(19):10706-14. doi: 10.1128/JVI.78.19.10706-10714.2004. J Virol. 2004. PMID: 15367637 Free PMC article.
-
Identification of specific HIV-1 reverse transcriptase contacts to the viral RNA:tRNA complex by mass spectrometry and a primary amine selective reagent.Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):15988-93. doi: 10.1073/pnas.252550199. Epub 2002 Dec 2. Proc Natl Acad Sci U S A. 2002. PMID: 12461175 Free PMC article.
-
The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription.J Virol. 2002 Mar;76(5):2329-39. doi: 10.1128/jvi.76.5.2329-2339.2002. J Virol. 2002. PMID: 11836411 Free PMC article.
References
-
- Telesnitsky A. and Goff,S.P. (1997) Reverse transcriptase and the generation of retroviral DNA. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Laboratory Press, Cold Spring Harbor, NY, pp. 121–160. - PubMed
-
- Arts E.J. and Le Grice,S. (1998) Interaction of retroviral reverse transcriptase with template–primer duplexes during replication. Prog. Nucleic Acids Res. Mol. Biol., 58, 335–394. - PubMed
-
- Isel C., Marquet,R., Keith,G., Ehresmann,C. and Ehresmann,B. (1993) Modified nucleotides of tRNALys3 modulate primer/template loop–loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem., 268, 25269–25272. - PubMed
-
- Isel C., Ehresmann,C., Keith,G., Ehresmann,B. and Marquet,R. (1995) Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNALys3 (template/primer). J. Mol. Biol., 247, 236–250. - PubMed

