Genome-wide detection of alternative splicing in expressed sequences of human genes
- PMID: 11433032
- PMCID: PMC55780
- DOI: 10.1093/nar/29.13.2850
Genome-wide detection of alternative splicing in expressed sequences of human genes
Abstract
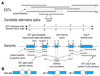
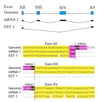
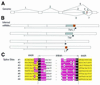
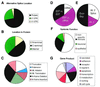
We have identified 6201 alternative splice relationships in human genes, through a genome-wide analysis of expressed sequence tags (ESTs). Starting with approximately 2.1 million human mRNA and EST sequences, we mapped expressed sequences onto the draft human genome sequence and only accepted splices that obeyed the standard splice site consensus. A large fraction (47%) of these were observed multiple times, indicating that they comprise a substantial fraction of the mRNA species. The vast majority of the detected alternative forms appear to be novel, and produce highly specific, biologically meaningful control of function in both known and novel human genes, e.g. specific removal of the lysosomal targeting signal from HLA-DM beta chain, replacement of the C-terminal transmembrane domain and cytoplasmic tail in an FC receptor beta chain homolog with a different transmembrane domain and cytoplasmic tail, likely modulating its signal transduction activity. Our data indicate that a large proportion of human genes, probably 42% or more, are alternatively spliced, but that this appears to be observed mainly in certain types of molecules (e.g. cell surface receptors) and systemic functions, particularly the immune system and nervous system. These results provide a comprehensive dataset for understanding the role of alternative splicing in the human genome, accessible at http://www.bioinformatics.ucla.edu/HASDB.
Figures

References
-
- Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. - PubMed
-
- Boise L.H., Gonzalez-Garcia,M., Postema,C.E., Ding,L., Lindsten,T., Turka,L.A., Mao,X., Nunez,G. and Thompson,C.B. (1993) bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell, 74, 597–608. - PubMed
-
- Fettiplace R. and Fuchs,P.A. (1999) Mechanisms of hair cell tuning. Annu. Rev. Physiol., 61, 809–834. - PubMed
-
- Schmucker D., Clemens,J.C., Shu,H., Worby,C.A., Xiao,J., Muda,M., Dixon,J.E. and Zipursky,S.L. (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell, 101, 671–684. - PubMed
-
- Smith C.W.J. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

