Phenotype microarrays for high-throughput phenotypic testing and assay of gene function
- PMID: 11435407
- PMCID: PMC311101
- DOI: 10.1101/gr.186501
Phenotype microarrays for high-throughput phenotypic testing and assay of gene function
Abstract
The bacterium Escherichia coli is used as a model cellular system to test and validate a new technology called Phenotype MicroArrays (PMs). PM technology is a high-throughput technology for simultaneous testing of a large number of cellular phenotypes. It consists of preconfigured well arrays in which each well tests a different cellular phenotype and an automated instrument that continuously monitors and records the response of the cells in all wells of the arrays. For example, nearly 700 phenotypes of E. coli can be assayed by merely pipetting a cell suspension into seven microplate arrays. PMs can be used to directly assay the effects of genetic changes on cells, especially gene knock-outs. Here, we provide data on phenotypic analysis of six strains and show that we can detect expected phenotypes as well as, in some cases, unexpected phenotypes.
Figures




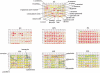

References
-
- Beck E, Ludwig G, Auerswald EA, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. - PubMed
-
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
-
- Bochner BR. New methods aid microbial identification. Bio/Technology. 1988;6:756.
-
- ————— “Breathprints” at the microbial level. ASM News. 1989a;55:536–539.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
