Second-site suppressors of Rous sarcoma virus Ca mutations: evidence for interdomain interactions
- PMID: 11435564
- PMCID: PMC114412
- DOI: 10.1128/JVI.75.15.6850-6856.2001
Second-site suppressors of Rous sarcoma virus Ca mutations: evidence for interdomain interactions
Abstract
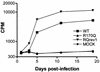
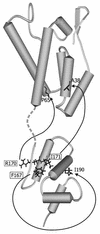
The capsid (CA) protein, the major structural component of retroviruses, forms a shell that encases the ribonucleoprotein complex in the virion core. The most conserved region of CA, approximately 20 amino acids of the major homology region (MHR), lies within the carboxy-terminal domain of the protein. Structural and sequence similarities among CA proteins of retroviruses and the CA-like proteins of hepatitis B virus and various retrotransposons suggest that the MHR is involved in an aspect of replication common to these reverse-transcribing elements. Conservative substitutions in this region of the Rous sarcoma virus protein were lethal due to a severe deficiency in reverse transcription, in spite of the presence of an intact genome and active reverse transcriptase in the particles. This finding suggests that the mutations interfered with normal interactions among these constituents. A total of four genetic suppressors of three lethal MHR mutations have now been identified. All four map to the sequence encoding the CA-spacer peptide (SP) region of Gag. The F167Y mutation in the MHR was fully suppressed by a single amino acid change in the alpha helix immediately downstream of the MHR, a region that forms the major dimer interface in human immunodeficiency virus CA. This finding suggests that the F167Y mutation indirectly interfered with dimerization. The F167Y defect could also be repaired by a second, independent suppressor in the C-terminal SP that was removed from CA during maturation. This single residue change, which increased the rate of SP cleavage, apparently corrected the F167Y defect by modifying the maturation pathway. More surprising was the isolation of suppressors of the R170Q and L171V MHR mutations, which mapped to the N-terminal domain of the CA protein. This finding suggests that the two domains, which in the monomeric protein are separated by a flexible linker, must communicate with each other at some unidentified point in the viral replication cycle.
Figures



References
-
- Campos-Olivas R, Newman J L, Summers M F. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

