Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur
- PMID: 11435581
- PMCID: PMC114429
- DOI: 10.1128/JVI.75.15.7009-7017.2001
Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur
Abstract
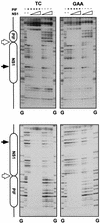
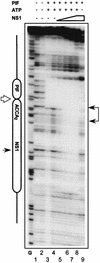
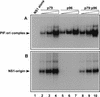
Parvoviral rolling hairpin replication generates palindromic genomic concatemers whose junctions are resolved to give unit-length genomes by a process involving DNA replication initiated at origins derived from each viral telomere. The left-end origin of minute virus of mice (MVM), oriL, contains binding sites for the viral initiator nickase, NS1, and parvovirus initiation factor (PIF), a member of the emerging KDWK family of transcription factors. oriL is generated as an active form, oriL(TC), and as an inactive form, oriL(GAA), which contains a single additional nucleotide inserted between the NS1 and PIF sites. Here we examined the interactions on oriL(TC) which lead to activation of NS1 by PIF. The two subunits of PIF, p79 and p96, cooperatively bind two ACGT half-sites, which can be flexibly spaced. When coexpressed from recombinant baculoviruses, the PIF subunits preferentially form heterodimers which, in the presence of ATP, show cooperative binding with NS1 on oriL, but this interaction is preferentially enhanced on oriL(TC) compared to oriL(GAA). Without ATP, NS1 is unable to bind stably to its cognate site, but PIF facilitates this interaction, rendering the NS1 binding site, but not the nick site, resistant to DNase I. Varying the spacing of the PIF half-sites shows that the distance between the NS1 binding site and the NS1-proximal half-site is critical for nickase activation, whereas the position of the distal half-site is unimportant. When expressed separately, both PIF subunits form homodimers that bind site specifically to oriL, but only complexes containing p79 activate the NS1 nickase function.
Figures

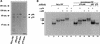



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

